QTL analysis of dietary obesity in C57BL/6byj X 129P3/J F2 mice: diet- and sex-dependent effects
- PMID: 23922663
- PMCID: PMC3726688
- DOI: 10.1371/journal.pone.0068776
QTL analysis of dietary obesity in C57BL/6byj X 129P3/J F2 mice: diet- and sex-dependent effects
Abstract
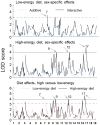
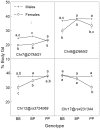
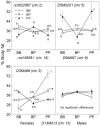
Obesity is a heritable trait caused by complex interactions between genes and environment, including diet. Gene-by-diet interactions are difficult to study in humans because the human diet is hard to control. Here, we used mice to study dietary obesity genes, by four methods. First, we bred 213 F2 mice from strains that are susceptible [C57BL/6ByJ (B6)] or resistant [129P3/J (129)] to dietary obesity. Percent body fat was assessed after mice ate low-energy diet and again after the same mice ate high-energy diet for 8 weeks. Linkage analyses identified QTLs associated with dietary obesity. Three methods were used to filter candidate genes within the QTL regions: (a) association mapping was conducted using >40 strains; (b) differential gene expression and (c) comparison of genomic DNA sequence, using two strains closely related to the progenitor strains from Experiment 1. The QTL effects depended on whether the mice were male or female or which diet they were recently fed. After feeding a low-energy diet, percent body fat was linked to chr 7 (LOD=3.42). After feeding a high-energy diet, percent body fat was linked to chr 9 (Obq5; LOD=3.88), chr 12 (Obq34; LOD=3.88), and chr 17 (LOD=4.56). The Chr 7 and 12 QTLs were sex dependent and all QTL were diet-dependent. The combination of filtering methods highlighted seven candidate genes within the QTL locus boundaries: Crx, Dmpk, Ahr, Mrpl28, Glo1, Tubb5, and Mut. However, these filtering methods have limitations so gene identification will require alternative strategies, such as the construction of congenics with very small donor regions.
Conflict of interest statement
Figures



References
-
- Stunkard AJ, Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, et al. (1986) An adoption study of human obesity. N Engl J Med 314: 193–198. - PubMed
-
- Sims EA, Horton ES (1968) Endocrine and metabolic adaptation to obesity and starvation. Am J Clin Nutr 21: 1455–1470. - PubMed
-
- Bouchard C, Tremblay A, Despres JP, Nadeau A, Lupien PJ, et al. (1990) The response to long-term overfeeding in identical twins. N Engl J Med 322: 1477–1482. - PubMed
-
- Fenton PF, Dowling MT (1953) Studies on obesity. I. Nutritional obesity in mice. J Nutr 49: 319–331. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

