An activating mutation reveals a second binding mode of the integrin α2 I domain to the GFOGER motif in collagens
- PMID: 23922814
- PMCID: PMC3726769
- DOI: 10.1371/journal.pone.0069833
An activating mutation reveals a second binding mode of the integrin α2 I domain to the GFOGER motif in collagens
Abstract
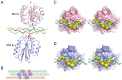
The GFOGER motif in collagens (O denotes hydroxyproline) represents a high-affinity binding site for all collagen-binding integrins. Other GxOGER motifs require integrin activation for maximal binding. The E318W mutant of the integrin α2β1 I domain displays a relaxed collagen specificity, typical of an active state. E318W binds more strongly than the wild-type α2 I domain to GMOGER, and forms a 2:1 complex with a homotrimeric, collagen-like, GFOGER peptide. Crystal structure analysis of this complex reveals two E318W I domains, A and B, bound to a single triple helix. The E318W I domains are virtually identical to the collagen-bound wild-type I domain, suggesting that the E318W mutation activates the I domain by destabilising the unligated conformation. E318W I domain A interacts with two collagen chains similarly to wild-type I domain (high-affinity mode). E318W I domain B makes favourable interactions with only one collagen chain (low-affinity mode). This observation suggests that single GxOGER motifs in the heterotrimeric collagens V and IX may support binding of activated integrins.
Conflict of interest statement
Figures


References
-
- Leitinger B (2011) Transmembrane collagen receptors. Annu Rev Cell Dev Biol 27: 265–290. - PubMed
-
- Lee JO, Rieu P, Arnaout MA, Liddington R (1995) Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18). Cell 80: 631–638. - PubMed
-
- Huang C, Springer TA (1995) A binding interface on the I domain of lymphocyte function-associated antigen-1 (LFA-1) required for specific interaction with intercellular adhesion molecule 1 (ICAM-1). J Biol Chem 270: 19008–19016. - PubMed
-
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

