A unique mitochondrial transcription factor B protein in Dictyostelium discoideum
- PMID: 23923009
- PMCID: PMC3724811
- DOI: 10.1371/journal.pone.0070614
A unique mitochondrial transcription factor B protein in Dictyostelium discoideum
Abstract
Unlike their bacteriophage homologs, mitochondrial RNA polymerases require the assistance of transcription factors in order to transcribe mitochondrial DNA efficiently. The transcription factor A family has been shown to be important for transcription of the human mitochondrial DNA, with some of its regulatory activity located in its extended C-terminal tail. The mitochondrial transcription factor B family often has functions not only in transcription, but also in mitochondrial rRNA modification, a hallmark of its α-proteobacterial origin. We have identified and characterised a mitochondrial transcription factor B homolog in the soil dwelling cellular slime mould Dictyostelium discoideum, an organism widely established as a model for studying eukaryotic cell biology. Using in bacterio functional assays, we demonstrate that the mitochondrial transcription factor B homolog not only functions as a mitochondrial transcription factor, but that it also has a role in rRNA methylation. Additionally, we show that the transcriptional activation properties of the D. discoideum protein are located in its extended C-terminal tail, a feature not seen before in the mitochondrial transcription factor B family, but reminiscent of the human mitochondrial transcription factor A. This report contributes to our current understanding of the complexities of mitochondrial transcription, and its evolution in eukaryotes.
Conflict of interest statement
Figures



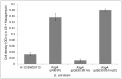
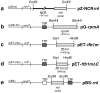

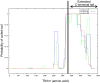

Similar articles
-
The Dictyostelium discoideum homologue of Twinkle, Twm1, is a mitochondrial DNA helicase, an active primase and promotes mitochondrial DNA replication.BMC Mol Biol. 2018 Dec 19;19(1):12. doi: 10.1186/s12867-018-0114-7. BMC Mol Biol. 2018. PMID: 30563453 Free PMC article.
-
Identification of the homolog of cell-counting factor in the cellular slime mold Dictyostelium discoideum.J Biochem Mol Biol Biophys. 2002 Oct;6(5):351-5. doi: 10.1080/1025814021000014001. J Biochem Mol Biol Biophys. 2002. PMID: 12385972
-
Characterization of a novel small RNA encoded by Dictyostelium discoideum mitochondrial DNA.Mol Gen Genet. 1998 Jan;257(2):124-31. doi: 10.1007/s004380050631. Mol Gen Genet. 1998. PMID: 9491070
-
Mitochondrial biology and disease in Dictyostelium.Int Rev Cytol. 2007;263:207-52. doi: 10.1016/S0074-7696(07)63005-8. Int Rev Cytol. 2007. PMID: 17725968 Review.
-
The Dictyostelium model for mitochondrial biology and disease.Int J Dev Biol. 2019;63(8-9-10):497-508. doi: 10.1387/ijdb.190233pf. Int J Dev Biol. 2019. PMID: 31840787 Review.
Cited by
-
Phylogenetic Reconstruction Shows Independent Evolutionary Origins of Mitochondrial Transcription Factors from an Ancient Family of RNA Methyltransferase Proteins.J Mol Evol. 2018 Jun;86(5):277-282. doi: 10.1007/s00239-018-9842-z. Epub 2018 Apr 25. J Mol Evol. 2018. PMID: 29691606 Free PMC article.
-
The Dictyostelium discoideum homologue of Twinkle, Twm1, is a mitochondrial DNA helicase, an active primase and promotes mitochondrial DNA replication.BMC Mol Biol. 2018 Dec 19;19(1):12. doi: 10.1186/s12867-018-0114-7. BMC Mol Biol. 2018. PMID: 30563453 Free PMC article.
-
A transcriptional factor B paralog functions as an activator to DNA damage-responsive expression in archaea.Nucleic Acids Res. 2018 Aug 21;46(14):7085-7096. doi: 10.1093/nar/gky236. Nucleic Acids Res. 2018. PMID: 29618058 Free PMC article.
References
-
- Gray MW, Lang BF, Burger G (2004) Mitochondria of protists. Annu Rev Genet 38: 477–524. - PubMed
-
- Asin-Cayuela J, Gustafsson CM (2007) Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci 32: 111–117. - PubMed
-
- Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW, et al. (1997) On the evolution of the single-subunit RNA polymerases. J Mol Evol 45: 671–681. - PubMed
-
- Shutt TE, Gray MW (2006) Homologs of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethyladenosine methyltransferase of the mitochondrial endosymbiont. Mol Biol Evol 23: 1169–1179. - PubMed
-
- Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

