MatrixCatch--a novel tool for the recognition of composite regulatory elements in promoters
- PMID: 23924163
- PMCID: PMC3754795
- DOI: 10.1186/1471-2105-14-241
MatrixCatch--a novel tool for the recognition of composite regulatory elements in promoters
Abstract
Background: Accurate recognition of regulatory elements in promoters is an essential prerequisite for understanding the mechanisms of gene regulation at the level of transcription. Composite regulatory elements represent a particular type of such transcriptional regulatory elements consisting of pairs of individual DNA motifs. In contrast to the present approach, most available recognition techniques are based purely on statistical evaluation of the occurrence of single motifs. Such methods are limited in application, since the accuracy of recognition is greatly dependent on the size and quality of the sequence dataset. Methods that exploit available knowledge and have broad applicability are evidently needed.
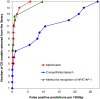
Results: We developed a novel method to identify composite regulatory elements in promoters using a library of known examples. In depth investigation of regularities encoded in known composite elements allowed us to introduce a new characteristic measure and to improve the specificity compared with other methods. Tests on an established benchmark and real genomic data show that our method outperforms other available methods based either on known examples or statistical evaluations. In addition to better recognition, a practical advantage of this method is first the ability to detect a high number of different types of composite elements, and second direct biological interpretation of the identified results. The program is available at http://gnaweb.helmholtz-hzi.de/cgi-bin/MCatch/MatrixCatch.pl and includes an option to extend the provided library by user supplied data.
Conclusions: The novel algorithm for the identification of composite regulatory elements presented in this paper was proved to be superior to existing methods. Its application to tissue specific promoters identified several highly specific composite elements with relevance to their biological function. This approach together with other methods will further advance the understanding of transcriptional regulation of genes.
Figures


References
-
- Waleev T, Shtokalo D, Konovalova T, Voss N, Cheremushkin E, Stegmaier P, Kel-Margoulis O, Wingender E, Kel A. Composite Module Analyst: identification of transcription factor binding site combinations using genetic algorithm. Nucleic Acids Res. 2006;34:W541–W545. doi: 10.1093/nar/gkl342. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

