Notch signaling represses p63 expression in the developing surface ectoderm
- PMID: 23924630
- PMCID: PMC3754476
- DOI: 10.1242/dev.093948
Notch signaling represses p63 expression in the developing surface ectoderm
Abstract
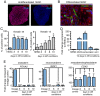
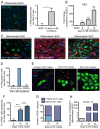
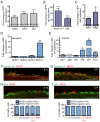
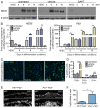
The development of the mature epidermis requires a coordinated sequence of signaling events and transcriptional changes to specify surface ectodermal progenitor cells to the keratinocyte lineage. The initial events that specify epidermal keratinocytes from ectodermal progenitor cells are not well understood. Here, we use both developing mouse embryos and human embryonic stem cells (hESCs) to explore the mechanisms that direct keratinocyte fate from ectodermal progenitor cells. We show that both hESCs and murine embryos express p63 before keratin 14. Furthermore, we find that Notch signaling is activated before p63 expression in ectodermal progenitor cells. Inhibition of Notch signaling pharmacologically or genetically reveals a negative regulatory role for Notch signaling in p63 expression during ectodermal specification in hESCs or mouse embryos, respectively. Taken together, these data reveal a role for Notch signaling in the molecular control of ectodermal progenitor cell specification to the epidermal keratinocyte lineage.
Keywords: Ectoderm; Keratinocyte specification; Notch signaling; p63.
Figures


References
-
- Aberdam E., Barak E., Rouleau M., de LaForest S., Berrih-Aknin S., Suter D. M., Krause K.-H., Amit M., Itskovitz-Eldor J., Aberdam D. (2008). A pure population of ectodermal cells derived from human embryonic stem cells. Stem Cells 26, 440–444 - PubMed
-
- Bernando A. S., Faial T., Gardner L., Niakan K. K., Ortmann D., Senner C. E., Callery E. M., Trotter M. W., Hemberger M., Smith J. C., et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155 - PMC - PubMed
-
- Bouras T., Pal B., Vaillant F., Harburg G., Asselin-Labat M.-L., Oakes S. R., Lindeman G. J., Visvader J. E. (2008). Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3, 429–441 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

