Direct assessment of transcription fidelity by high-resolution RNA sequencing
- PMID: 23925128
- PMCID: PMC3799451
- DOI: 10.1093/nar/gkt698
Direct assessment of transcription fidelity by high-resolution RNA sequencing
Abstract
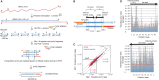
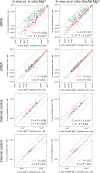
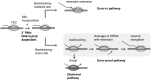
Cancerous and aging cells have long been thought to be impacted by transcription errors that cause genetic and epigenetic changes. Until now, a lack of methodology for directly assessing such errors hindered evaluation of their impact to the cells. We report a high-resolution Illumina RNA-seq method that can assess noncoded base substitutions in mRNA at 10(-4)-10(-5) per base frequencies in vitro and in vivo. Statistically reliable detection of changes in transcription fidelity through ∼10(3) nt DNA sites assures that the RNA-seq can analyze the fidelity in a large number of the sites where errors occur. A combination of the RNA-seq and biochemical analyses of the positions for the errors revealed two sequence-specific mechanisms that increase transcription fidelity by Escherichia coli RNA polymerase: (i) enhanced suppression of nucleotide misincorporation that improves selectivity for the cognate substrate, and (ii) increased backtracking of the RNA polymerase that decreases a chance of error propagation to the full-length transcript after misincorporation and provides an opportunity to proofread the error. This method is adoptable to a genome-wide assessment of transcription fidelity.
Figures




References
-
- Paoloni-Giacobino A, Rossier C, Papasavvas MP, Antonarakis SE. Frequency of replication/transcription errors in (A)/(T) runs of human genes. Hum. Genet. 2001;109:40–47. - PubMed
-
- Rodin SN, Rodin AS, Juhasz A, Holmquist GP. Cancerous hyper-mutagenesis in p53 genes is possibly associated with transcriptional bypass of DNA lesions. Mutat. Res. 2002;510:153–168. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

