PPR proteins shed a new light on RNase P biology
- PMID: 23925311
- PMCID: PMC3858429
- DOI: 10.4161/rna.25273
PPR proteins shed a new light on RNase P biology
Abstract
A fast growing number of studies identify pentatricopeptide repeat (PPR) proteins as major players in gene expression processes. Among them, a subset of PPR proteins called PRORP possesses RNase P activity in several eukaryotes, both in nuclei and organelles. RNase P is the endonucleolytic activity that removes 5' leader sequences from tRNA precursors and is thus essential for translation. Before the characterization of PRORP, RNase P enzymes were thought to occur universally as ribonucleoproteins, although some evidence implied that some eukaryotes or cellular compartments did not use RNA for RNase P activity. The characterization of PRORP reveals a two-domain enzyme, with an N-terminal domain containing multiple PPR motifs and assumed to achieve target specificity and a C-terminal domain holding catalytic activity. The nature of PRORP interactions with tRNAs suggests that ribonucleoprotein and protein-only RNase P enzymes share a similar substrate binding process.
Keywords: RNA binding protein; RNase P; evolution; pentatricopeptide repeat; tRNA maturation.
Figures
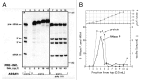
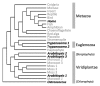

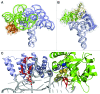
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
