Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells
- PMID: 23926201
- PMCID: PMC6002776
- DOI: 10.1126/scitranslmed.3006034
Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells
Abstract
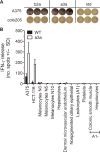
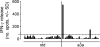
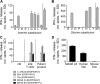
MAGE A3, which belongs to the family of cancer-testis antigens, is an attractive target for adoptive therapy given its reactivation in various tumors and limited expression in normal tissues. We developed an affinity-enhanced T cell receptor (TCR) directed to a human leukocyte antigen (HLA)-A*01-restricted MAGE A3 antigen (EVDPIGHLY) for use in adoptive therapy. Extensive preclinical investigations revealed no off-target antigen recognition concerns; nonetheless, administration to patients of T cells expressing the affinity-enhanced MAGE A3 TCR resulted in a serious adverse event (SAE) and fatal toxicity against cardiac tissue. We present a description of the preclinical in vitro functional analysis of the MAGE A3 TCR, which failed to reveal any evidence of off-target activity, and a full analysis of the post-SAE in vitro investigations, which reveal cross-recognition of an off-target peptide. Using an amino acid scanning approach, a peptide from the muscle protein Titin (ESDPIVAQY) was identified as an alternative target for the MAGE A3 TCR and the most likely cause of in vivo toxicity. These results demonstrate that affinity-enhanced TCRs have considerable effector functions in vivo and highlight the potential safety concerns for TCR-engineered T cells. Strategies such as peptide scanning and the use of more complex cell cultures are recommended in preclinical studies to mitigate the risk of off-target toxicity in future clinical investigations.
Conflict of interest statement
Figures




References
-
- Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, Davis JL, Morgan RA, Merino MJ, Sherry RM, Hughes MS, Kammula US, Phan GQ, Lim RM, Wank SA, Restifo NP, Robbins PF, Laurencot CM, Rosenberg SA. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19:620–626. - PMC - PubMed
-
- Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee CC, Levy CL, Li YF, El-Gamil M, Schwarz SL, Laurencot C, Rosenberg SA. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–924. - PMC - PubMed
-
- Kalos M, Rapoport AP, Stadtmauer EA, Vogl DT, Weiss BM, Binder-Scholl GK, Smethurst DP, Kulikovskaya I, Gupta M, Macatee TL, Finklestein JM, Brewer JE, Bennett AD, Gerry AB, Pumphrey NJ, Tayton-Martin HK, Veloso E, Zheng Z, Badros AZ, Yanovich S, Akpek G, McConville H, Philip S, Levine BL, Jakobsen BK, June CH. Prolonged T cell persistence, homing to marrow and selective targeting of antigen positive tumor in multiple myeloma patients following adoptive transfer of T cells genetically engineered to express an affinity-enhanced T cell receptor against the cancer testis antigens NY-ESO-1 and Lage-1. Blood (ASH Annu Meet Abstr) 2012;120:755.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

