Sustained knockdown of a disease-causing gene in patient-specific induced pluripotent stem cells using lentiviral vector-based gene therapy
- PMID: 23926210
- PMCID: PMC3754465
- DOI: 10.5966/sctm.2013-0017
Sustained knockdown of a disease-causing gene in patient-specific induced pluripotent stem cells using lentiviral vector-based gene therapy
Abstract
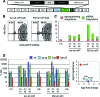
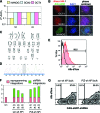
Patient-specific induced pluripotent stem cells (iPSCs) hold great promise for studies on disease-related developmental processes and may serve as an autologous cell source for future treatment of many hereditary diseases. New genetic engineering tools such as zinc finger nucleases and transcription activator-like effector nuclease allow targeted correction of monogenetic disorders but are very cumbersome to establish. Aiming at studies on the knockdown of a disease-causing gene, lentiviral vector-mediated expression of short hairpin RNAs (shRNAs) is a valuable option, but it is limited by silencing of the knockdown construct upon epigenetic remodeling during differentiation. Here, we propose an approach for the expression of a therapeutic shRNA in disease-specific iPSCs using third-generation lentiviral vectors. Targeting severe α-1-antitrypsin (A1AT) deficiency, we overexpressed a human microRNA 30 (miR30)-styled shRNA directed against the PiZ variant of A1AT, which is known to cause chronic liver damage in affected patients. This knockdown cassette is traceable from clonal iPSC lines to differentiated hepatic progeny via an enhanced green fluorescence protein reporter expressed from the same RNA-polymerase II promoter. Importantly, the cytomegalovirus i/e enhancer chicken β actin (CAG) promoter-driven expression of this construct is sustained without transgene silencing during hepatic differentiation in vitro and in vivo. At low lentiviral copy numbers per genome we confirmed a functional relevant reduction (-66%) of intracellular PiZ protein in hepatic cells after differentiation of patient-specific iPSCs. In conclusion, we have demonstrated that lentiviral vector-mediated expression of shRNAs can be efficiently used to knock down and functionally evaluate disease-related genes in patient-specific iPSCs.
Keywords: Gene therapy; Hepatocyte differentiation; Lentiviral vector; Liver; Pluripotent stem cells; RNAi.
Figures




References
-
- Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: Basic to translational. Biochem J. 2012;443:603–618. - PubMed
-
- Frimpong K, Spector SA. Cotransduction of nondividing cells using lentiviral vectors. Gene Ther. 2000;7:1562–1569. - PubMed
-
- Yu X, Zhan X, D'Costa J, et al. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol Ther. 2003;7:827–838. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

