Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease
- PMID: 23926354
- PMCID: PMC3807292
- DOI: 10.1128/JVI.01123-13
Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease
Abstract
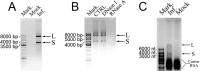
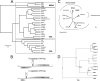
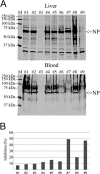
Boid inclusion body disease (BIBD) is a progressive, usually fatal disease of constrictor snakes, characterized by cytoplasmic inclusion bodies (IB) in a wide range of cell types. To identify the causative agent of the disease, we established cell cultures from BIBD-positive and -negative boa constrictors. The IB phenotype was maintained in cultured cells of affected animals, and supernatants from these cultures caused the phenotype in cultures originating from BIBD-negative snakes. Viruses were purified from the supernatants by ultracentrifugation and subsequently identified as arenaviruses. Purified virus also induced the IB phenotype in naive cells, which fulfilled Koch's postulates in vitro. One isolate, tentatively designated University of Helsinki virus (UHV), was studied in depth. Sequencing confirmed that UHV is a novel arenavirus species that is distinct from other known arenaviruses including those recently identified in snakes with BIBD. The morphology of UHV was established by cryoelectron tomography and subtomographic averaging, revealing the trimeric arenavirus spike structure at 3.2-nm resolution. Immunofluorescence, immunohistochemistry, and immunoblotting with a polyclonal rabbit antiserum against UHV and reverse transcription-PCR (RT-PCR) revealed the presence of genetically diverse arenaviruses in a large cohort of snakes with BIBD, confirming the causative role of arenaviruses. Some snakes were also found to carry arenavirus antibodies. Furthermore, mammalian cells (Vero E6) were productively infected with UHV, demonstrating the potential of arenaviruses to cross species barriers. In conclusion, we propose the newly identified lineage of arenaviruses associated with BIBD as a novel taxonomic entity, boid inclusion body disease-associated arenaviruses (BIBDAV), in the family Arenaviridae.
Figures






Comment in
-
Updated phylogenetic analysis of arenaviruses detected in boid snakes.J Virol. 2014 Jan;88(2):1399-400. doi: 10.1128/JVI.02753-13. J Virol. 2014. PMID: 24379418 Free PMC article. No abstract available.
-
Reply to "Updated phylogenetic analysis of arenaviruses detected in boid snakes".J Virol. 2014 Jan;88(2):1401. doi: 10.1128/JVI.03044-13. J Virol. 2014. PMID: 24379419 Free PMC article. No abstract available.
References
-
- Vidal N, Hedges SB. 2009. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C. R. Biol. 332:129–139. - PubMed
-
- Schumacher J, Jacobson ER, Homer BL, Gaskin JM. 1994. Inclusion body disease in boid snakes. J. Zoo Wildl. Med. 25:511–524.
-
- Carlisle-Nowak MS, Sullivan N, Carrigan M, Knight C, Ryan C, Jacobson ER. 1998. Inclusion body disease in two captive Australian pythons (Morelia spilota variegata and Morelia spilota spilota). Aust. Vet. J. 76:98–100. - PubMed
-
- Raymond JT, Garner MM, Nordhausen RW, Jacobson ER. 2001. A disease resembling inclusion body disease of boid snakes in captive palm vipers (Bothriechis marchi). J. Vet. Diagn. Invest. 13:82–86. - PubMed
-
- Fleming GJ, Heard DJ, Jacobson ER. 2003. Cytoplasmic inclusions in corn snakes, Elaphe guttata, resembling inclusion body disease of boid snakes. J. Herpetol. Med. Surg. 13:18–22.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

