Bacterial Swarming: A Model System for Studying Dynamic Self-assembly
- PMID: 23926448
- PMCID: PMC3733279
- DOI: 10.1039/B812146J
Bacterial Swarming: A Model System for Studying Dynamic Self-assembly
Abstract
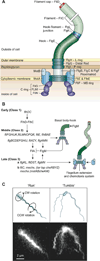
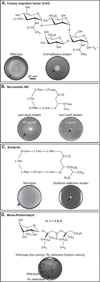
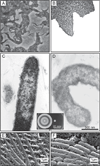
Bacterial swarming is an example of dynamic self-assembly in microbiology in which the collective interaction of a population of bacterial cells leads to emergent behavior. Swarming occurs when cells interact with surfaces, reprogram their physiology and behavior, and adapt to changes in their environment by coordinating their growth and motility with other cells in the colony. This review summarizes the salient biological and biophysical features of this system and describes our current understanding of swarming motility. We have organized this review into four sections: 1) The biophysics and mechanisms of bacterial motility in fluids and its relevance to swarming. 2) The role of cell/molecule, cell/surface, and cell/cell interactions during swarming. 3) The changes in physiology and behavior that accompany swarming motility. 4) A concluding discussion of several interesting, unanswered questions that is particularly relevant to soft matter scientists.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

