Single-cell semiconductor sequencing
- PMID: 23929110
- PMCID: PMC4024476
- DOI: 10.1007/978-1-62703-556-9_18
Single-cell semiconductor sequencing
Abstract
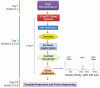
RNA-seq or transcriptome analysis of individual cells and small-cell populations is essential for virtually any biomedical field. It is especially critical for developmental, aging, and cancer biology as well as neuroscience where the enormous heterogeneity of cells present a significant methodological and conceptual challenge. Here we present two methods that allow for fast and cost-efficient transcriptome sequencing from ultra-small amounts of tissue or even from individual cells using semiconductor sequencing technology (Ion Torrent, Life Technologies). The first method is a reduced representation sequencing which maximizes capture of RNAs and preserves transcripts' directionality. The second, a template-switch protocol, is designed for small mammalian neurons. Both protocols, from cell/tissue isolation to final sequence data, take up to 4 days. The efficiency of these protocols has been validated with single hippocampal neurons and various invertebrate tissues including individually identified neurons within a simpler memory-forming circuit of Aplysia californica and early (1-, 2-, 4-, 8-cells) embryonic and developmental stages from basal metazoans.
Figures






References
-
- Moroz LL, Kohn AB. Single-neuron transcriptome and methylome sequencing for epigenomic analysis of aging. In: Tollefsbol Trygve O., editor. Biological Aging: Methods and Protocols, Methods in Molecular Biology: Methods and Protocols. Vol. 1048. # Springer Science +Business Media; New York: 2013. DOI 10.1007/978-1-62703-556-9_21. 2013. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

