Super-resolution fluorescence microscopy of the cardiac connexome reveals plakophilin-2 inside the connexin43 plaque
- PMID: 23929525
- PMCID: PMC3797628
- DOI: 10.1093/cvr/cvt191
Super-resolution fluorescence microscopy of the cardiac connexome reveals plakophilin-2 inside the connexin43 plaque
Abstract
Aims: Cell function requires formation of molecular clusters localized to discrete subdomains. The composition of these interactomes, and their spatial organization, cannot be discerned by conventional microscopy given the resolution constraints imposed by the diffraction limit of light (∼200-300 nm). Our aims were (i) Implement single-molecule imaging and analysis tools to resolve the nano-scale architecture of cardiac myocytes. (ii) Using these tools, to map two molecules classically defined as components 'of the desmosome' and 'of the gap junction', and defined their spatial organization.
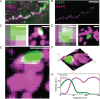
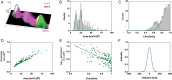
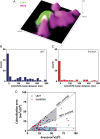
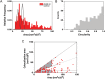
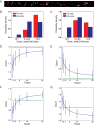
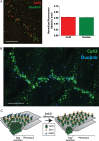
Methods and results: We built a set-up on a conventional inverted microscope using commercially available optics. Laser illumination, reducing, and oxygen scavenging conditions were used to manipulate the blinking behaviour of individual fluorescent reporters. Movies of blinking fluorophores were reconstructed to generate subdiffraction images at ∼20 nm resolution. With this method, we characterized clusters of connexin43 (Cx43) and of 'the desmosomal protein' plakophilin-2 (PKP2). In about half of Cx43 clusters, we observed overlay of Cx43 and PKP2 at the Cx43 plaque edge. SiRNA-mediated loss of Ankyrin-G expression yielded larger Cx43 clusters, of less regular shape, and larger Cx43-PKP2 subdomains. The Cx43-PKP2 subdomain was validated by a proximity ligation assay (PLA) and by Monte-Carlo simulations indicating an attraction between PKP2 and Cx43.
Conclusions: (i) Super-resolution fluorescence microscopy, complemented with Monte-Carlo simulations and PLAs, allows the study of the nanoscale organization of an interactome in cardiomyocytes. (ii) PKP2 and Cx43 share a common hub that permits direct physical interaction. Its relevance to excitability, electrical coupling, and arrhythmogenic right ventricular cardiomyopathy, is discussed.
Keywords: Ankyrin-G; Connexin43; Plakophilin-2.
Figures
References
-
- Born MW, E. Principles of Optics: Electromagnetic Theory of Propagation, Interference and Diffraction of Light. Cambridge: Cambridge University Press; 1997.
-
- Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi:10.1002/anie.200802376. - DOI - PubMed
-
- Baddeley D, Jayasinghe ID, Lam L, Rossberger S, Cannell MB, Soeller C. Optical single-channel resolution imaging of the ryanodine receptor distribution in rat cardiac myocytes. Proc Natl Acad Sci U S A. 2009;106:22275–22280. doi:10.1073/pnas.0908971106. - DOI - PMC - PubMed
-
- Soeller C, Baddeley D. Super-resolution imaging of EC coupling protein distribution in the heart. J Mol Cell Cardiol. 2013;58:32–40. doi:10.1016/j.yjmcc.2012.11.004. - DOI - PubMed
-
- Wong J, Baddeley D, Bushong EA, Yu Z, Ellisman MH, Hoshijima M, et al. Nanoscale distribution of ryanodine receptors and caveolin-3 in mouse ventricular myocytes: dilation of T-tubules near junctions. Biophys J. 2013;104:L22–L24. doi:10.1016/j.bpj.2013.02.059. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

