Splicing biomarkers of disease severity in myotonic dystrophy
- PMID: 23929620
- PMCID: PMC4099006
- DOI: 10.1002/ana.23992
Splicing biomarkers of disease severity in myotonic dystrophy
Abstract
Objective: To develop RNA splicing biomarkers of disease severity and therapeutic response in myotonic dystrophy type 1 (DM1) and type 2 (DM2).
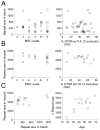
Methods: In a discovery cohort, we used microarrays to perform global analysis of alternative splicing in DM1 and DM2. The newly identified splicing changes were combined with previous data to create a panel of 50 putative splicing defects. In a validation cohort of 50 DM1 subjects, we measured the strength of ankle dorsiflexion (ADF) and then obtained a needle biopsy of tibialis anterior (TA) to analyze splice events in muscle RNA. The specificity of DM-associated splicing defects was assessed in disease controls. The CTG expansion size in muscle tissue was determined by Southern blot. The reversibility of splicing defects was assessed in transgenic mice by using antisense oligonucleotides to reduce levels of toxic RNA.
Results: Forty-two splicing defects were confirmed in TA muscle in the validation cohort. Among these, 20 events showed graded changes that correlated with ADF weakness. Five other splice events were strongly affected in DM1 subjects with normal ADF strength. Comparison to disease controls and mouse models indicated that splicing changes were DM-specific, mainly attributable to MBNL1 sequestration, and reversible in mice by targeted knockdown of toxic RNA. Splicing defects and weakness were not correlated with CTG expansion size in muscle tissue.
Interpretation: Alternative splicing changes in skeletal muscle may serve as biomarkers of disease severity and therapeutic response in myotonic dystrophy.
© 2013 American Neurological Association.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


References
-
- Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
-
- Liquori CL, Ricker K, Moseley ML, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–7. - PubMed
-
- Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. - PubMed
-
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–41. - PubMed
-
- Orengo JP, Cooper TA. Alternative splicing in disease. Adv Exp Med Biol. 2007;623:212–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K23 AR055947/AR/NIAMS NIH HHS/United States
- UL1RR024160-06S4/RR/NCRR NIH HHS/United States
- K08 NS064293/NS/NINDS NIH HHS/United States
- AR48143/AR/NIAMS NIH HHS/United States
- UL1 RR024160/RR/NCRR NIH HHS/United States
- K08NS064293/NS/NINDS NIH HHS/United States
- U01 NS072323/NS/NINDS NIH HHS/United States
- AR049077/AR/NIAMS NIH HHS/United States
- NS048843/NS/NINDS NIH HHS/United States
- AR055947/AR/NIAMS NIH HHS/United States
- R01 AR049077/AR/NIAMS NIH HHS/United States
- UL1 TR000042/TR/NCATS NIH HHS/United States
- P50 NS048843/NS/NINDS NIH HHS/United States
- R01 AR046799/AR/NIAMS NIH HHS/United States
- UL1RR024160/RR/NCRR NIH HHS/United States
- P01 NS058901/NS/NINDS NIH HHS/United States
- K24 AR048143/AR/NIAMS NIH HHS/United States
- U54 NS048843/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

