Spatial dynamics of chromosome translocations in living cells
- PMID: 23929981
- PMCID: PMC6324928
- DOI: 10.1126/science.1237150
Spatial dynamics of chromosome translocations in living cells
Abstract
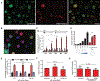
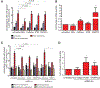
Chromosome translocations are a hallmark of cancer cells. We have developed an experimental system to visualize the formation of translocations in living cells and apply it to characterize the spatial and dynamic properties of translocation formation. We demonstrate that translocations form within hours of the occurrence of double-strand breaks (DSBs) and that their formation is cell cycle-independent. Translocations form preferentially between prepositioned genome elements, and perturbation of key factors of the DNA repair machinery uncouples DSB pairing from translocation formation. These observations generate a spatiotemporal framework for the formation of translocations in living cells.
Figures


References
-
- Mitelman F, Johansson B, Mertens F, Nat. Rev. Cancer 7, 233–245 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

