Negatively charged metal oxide nanoparticles interact with the 20S proteasome and differentially modulate its biologic functional effects
- PMID: 23930940
- PMCID: PMC3946455
- DOI: 10.1021/nn402416h
Negatively charged metal oxide nanoparticles interact with the 20S proteasome and differentially modulate its biologic functional effects
Abstract
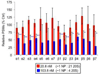
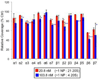
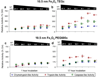
The multicatalytic ubiquitin-proteasome system (UPS) carries out proteolysis in a highly orchestrated way and regulates a large number of cellular processes. Deregulation of the UPS in many disorders has been documented. In some cases, such as carcinogenesis, elevated proteasome activity has been implicated in disease development, while the etiology of other diseases, such as neurodegeneration, includes decreased UPS activity. Therefore, agents that alter proteasome activity could suppress as well as enhance a multitude of diseases. Metal oxide nanoparticles, often developed as diagnostic tools, have not previously been tested as modulators of proteasome activity. Here, several types of metal oxide nanoparticles were found to adsorb to the proteasome and show variable preferential binding for particular proteasome subunits with several peptide binding "hotspots" possible. These interactions depend on the size, charge, and concentration of the nanoparticles and affect proteasome activity in a time-dependent manner. Should metal oxide nanoparticles increase proteasome activity in cells, as they do in vitro, unintended effects related to changes in proteasome function can be expected.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
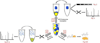



References
-
- Glickman MH, Ciechanover A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002;82:373–428. - PubMed
-
- Voges D, Zwickl P, Baumeister W. The 26s Proteasome: A Molecular Machine Designed for Controlled Proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. - PubMed
-
- Scheffner M, Nuber U, Huibregtse JM. Protein Ubiquitination Involving an E1-E2-E3 Enzyme Ubiquitin Thioester Cascade. Nature. 1995;373:81–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

