Conservation of protein structure over four billion years
- PMID: 23932589
- PMCID: PMC3774310
- DOI: 10.1016/j.str.2013.06.020
Conservation of protein structure over four billion years
Abstract
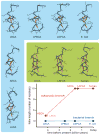
Little is known about the evolution of protein structures and the degree of protein structure conservation over planetary time scales. Here, we report the X-ray crystal structures of seven laboratory resurrections of Precambrian thioredoxins dating up to approximately four billion years ago. Despite considerable sequence differences compared with extant enzymes, the ancestral proteins display the canonical thioredoxin fold, whereas only small structural changes have occurred over four billion years. This remarkable degree of structure conservation since a time near the last common ancestor of life supports a punctuated-equilibrium model of structure evolution in which the generation of new folds occurs over comparatively short periods and is followed by long periods of structural stasis.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Benner SA, Sassi SO, Gaucher EA. Molecular paleoscience: systems biology from the past. Advances in enzymology and related areas of molecular biology. 2007;75:1–132. xi. - PubMed
-
- Caetano-Anolles G, Wang M, Caetano-Anolles D, Mittenthal JE. The origin, evolution and structure of the protein world. Biochem J. 2009;417:621–637. - PubMed
-
- Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography. 1994;50:760–763. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

