Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription
- PMID: 23932714
- PMCID: PMC3779836
- DOI: 10.1016/j.molcel.2013.07.010
Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription
Abstract
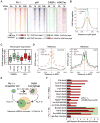
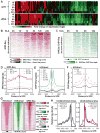
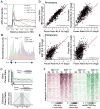
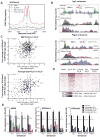
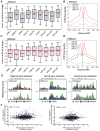
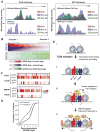
Recent studies suggest a hierarchical model in which lineage-determining factors act in a collaborative manner to select and prime cell-specific enhancers, thereby enabling signal-dependent transcription factors to bind and function in a cell-type-specific manner. Consistent with this model, TLR4 signaling primarily regulates macrophage gene expression through a pre-existing enhancer landscape. However, TLR4 signaling also induces priming of ∼3,000 enhancer-like regions de novo, enabling visualization of intermediates in enhancer selection and activation. Unexpectedly, we find that enhancer transcription precedes local mono- and dimethylation of histone H3 lysine 4 (H3K4me1/2). H3K4 methylation at de novo enhancers is primarily dependent on the histone methyltransferases Mll1, Mll2/4, and Mll3 and is significantly reduced by inhibition of RNA polymerase II elongation. Collectively, these findings suggest an essential role of enhancer transcription in H3K4me1/2 deposition at de novo enhancers that is independent of potential functions of the resulting eRNA transcripts.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

References
-
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annual review of pharmacology and toxicology. 2010;50:259–293. - PubMed
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genetics. 2012;44:148–156. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

