The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy
- PMID: 23933751
- PMCID: PMC3827746
- DOI: 10.1038/nn.3489
The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy
Abstract
Compelling evidence indicates that two autosomal recessive Parkinson's disease genes, PINK1 (PARK6) and Parkin (PARK2), cooperate to mediate the autophagic clearance of damaged mitochondria (mitophagy). Mutations in the F-box domain-containing protein Fbxo7 (encoded by PARK15) also cause early-onset autosomal recessive Parkinson's disease, by an unknown mechanism. Here we show that Fbxo7 participates in mitochondrial maintenance through direct interaction with PINK1 and Parkin and acts in Parkin-mediated mitophagy. Cells with reduced Fbxo7 expression showed deficiencies in translocation of Parkin to mitochondria, ubiquitination of mitofusin 1 and mitophagy. In Drosophila, ectopic overexpression of Fbxo7 rescued loss of Parkin, supporting a functional relationship between the two proteins. Parkinson's disease-causing mutations in Fbxo7 interfered with this process, emphasizing the importance of mitochondrial dysfunction in Parkinson's disease pathogenesis.
Figures

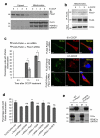
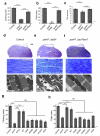

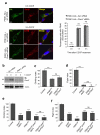

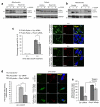
References
-
- Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0701075/MRC_/Medical Research Council/United Kingdom
- G1001253/MRC_/Medical Research Council/United Kingdom
- 089698/WT_/Wellcome Trust/United Kingdom
- H-1006/PUK_/Parkinson's UK/United Kingdom
- G0700183/MRC_/Medical Research Council/United Kingdom
- MR/J004758/1/MRC_/Medical Research Council/United Kingdom
- 309742/ERC_/European Research Council/International
- F-1002/PUK_/Parkinson's UK/United Kingdom
- G108/638/MRC_/Medical Research Council/United Kingdom
- MC_G1000735/MRC_/Medical Research Council/United Kingdom
- BB/F012764/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J007846/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- GR077544AIA/WT_/Wellcome Trust/United Kingdom
- G0802760/MRC_/Medical Research Council/United Kingdom
- G070091/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

