Evolutionary implications of a third lymphocyte lineage in lampreys
- PMID: 23934109
- PMCID: PMC3901013
- DOI: 10.1038/nature12467
Evolutionary implications of a third lymphocyte lineage in lampreys
Abstract
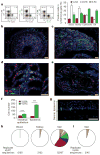
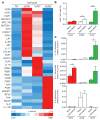
Jawed vertebrates (gnathostomes) and jawless vertebrates (cyclostomes) have different adaptive immune systems. Gnathostomes use T- and B-cell antigen receptors belonging to the immunoglobulin superfamily. Cyclostomes, the lampreys and hagfish, instead use leucine-rich repeat proteins to construct variable lymphocyte receptors (VLRs), two types of which, VLRA and VLRB, are reciprocally expressed by lymphocytes resembling gnathostome T and B cells. Here we define another lineage of T-cell-like lymphocytes that express the recently identified VLRC receptors. Both VLRC(+) and VLRA(+) lymphocytes express orthologues of genes that gnathostome γδ and αβ T cells use for their differentiation, undergo VLRC and VLRA assembly and repertoire diversification in the 'thymoid' gill region, and express their VLRs solely as cell-surface proteins. Our findings suggest that the genetic programmes for two primordial T-cell lineages and a prototypic B-cell lineage were already present in the last common vertebrate ancestor approximately 500 million years ago. We propose that functional specialization of distinct T-cell-like lineages was an ancient feature of a primordial immune system.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

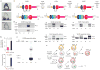
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

