High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity
- PMID: 23934178
- PMCID: PMC3782611
- DOI: 10.1038/nbt.2673
High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity
Abstract
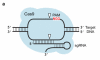
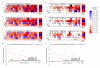
The RNA-programmable Cas9 endonuclease cleaves double-stranded DNA at sites complementary to a 20-base-pair guide RNA. The Cas9 system has been used to modify genomes in multiple cells and organisms, demonstrating its potential as a facile genome-engineering tool. We used in vitro selection and high-throughput sequencing to determine the propensity of eight guide-RNA:Cas9 complexes to cleave each of 10(12) potential off-target DNA sequences. The selection results predicted five off-target sites in the human genome that were confirmed to undergo genome cleavage in HEK293T cells upon expression of one of two guide-RNA:Cas9 complexes. In contrast to previous models, our results show that guide-RNA:Cas9 specificity extends past a 7- to 12-base-pair seed sequence. Our results also suggest a tradeoff between activity and specificity both in vitro and in cells as a shorter, less-active guide RNA is more specific than a longer, more-active guide RNA. High concentrations of guide-RNA:Cas9 complexes can cleave off-target sites containing mutations near or within the PAM that are not cleaved when enzyme concentrations are limiting.
Figures

Comment in
-
Staying on target with CRISPR-Cas.Nat Biotechnol. 2013 Sep;31(9):807-9. doi: 10.1038/nbt.2684. Nat Biotechnol. 2013. PMID: 24022156 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

