Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape
- PMID: 23934658
- PMCID: PMC3759695
- DOI: 10.1101/gad.223834.113
Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape
Abstract
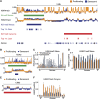
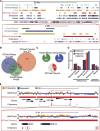
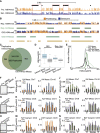
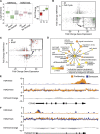
Senescence is a stable proliferation arrest, associated with an altered secretory pathway, thought to promote tumor suppression and tissue aging. While chromatin regulation and lamin B1 down-regulation have been implicated as senescence effectors, functional interactions between them are poorly understood. We compared genome-wide Lys4 trimethylation on histone H3 (H3K4me3) and H3K27me3 distributions between proliferating and senescent human cells and found dramatic differences in senescence, including large-scale domains of H3K4me3- and H3K27me3-enriched "mesas" and H3K27me3-depleted "canyons." Mesas form at lamin B1-associated domains (LADs) in replicative senescence and oncogene-induced senescence and overlap DNA hypomethylation regions in cancer, suggesting that pre-malignant senescent chromatin changes foreshadow epigenetic cancer changes. Hutchinson-Gilford progeria syndrome fibroblasts (mutant lamin A) also show evidence of H3K4me3 mesas, suggesting a link between premature chromatin changes and accelerated cell senescence. Canyons mostly form between LADs and are enriched in genes and enhancers. H3K27me3 loss is correlated with up-regulation of key senescence genes, indicating a link between global chromatin changes and local gene expression regulation. Lamin B1 reduction in proliferating cells triggers senescence and formation of mesas and canyons. Our data illustrate profound chromatin reorganization during senescence and suggest that lamin B1 down-regulation in senescence is a key trigger of global and local chromatin changes that impact gene expression, aging, and cancer.
Keywords: chromatin; gene expression; lamin B1; senescence.
Figures

References
-
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. 2008. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133: 1006–1018 - PubMed
-
- Adams PD 2009. Healing and hurting: Molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 36: 2–14 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
