Expression plasmids for use in Candida glabrata
- PMID: 23934995
- PMCID: PMC3789792
- DOI: 10.1534/g3.113.006908
Expression plasmids for use in Candida glabrata
Erratum in
- G3 (Bethesda). 2014 Jul;4(7):1361
Abstract
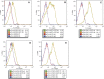
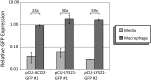
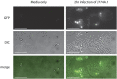
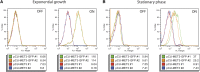
We describe a series of CEN/ARS episomal plasmids containing different Candida glabrata promoters, allowing for a range of constitutive or regulated expression of proteins in C. glabrata. The set of promoters includes three constitutive promoters (EGD2pr, HHT2pr, PDC1pr), two macrophage/phagocytosis-induced promoters (ACO2pr, LYS21pr), and one nutritionally regulated promoter (MET3pr). Each promoter was cloned into two plasmid backbones that differ in their selectable marker, URA3, or the dominant-selectable NAT1 gene, which confers resistance to the drug nourseothricin. Expression from the 12 resulting plasmids was assessed using GFP as a reporter and flow cytometry or quantitative reverse-transcription polymerase chain reaction to assess expression levels. Together this set of plasmids expands the toolkit of expression vectors available for use with C. glabrata.
Keywords: Candida glabrata; MET3; expression vector; inducible; macrophage.
Figures


References
-
- Adams C., Haldar D., Kamakaka R. T., 2005. Construction and characterization of a series of vectors for Schizosaccharomyces pombe. Yeast 22: 1307–1314 - PubMed
-
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
-
- Care R. S., Trevethick J., Binley K. M., Sudbery P. E., 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34: 792–798 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
