The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid
- PMID: 23935049
- PMCID: PMC3807434
- DOI: 10.1128/JB.00707-13
The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid
Abstract
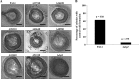
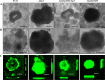
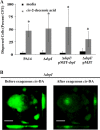
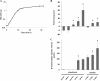
In the present study, we report the identification of a putative enoyl-coenzyme A (CoA) hydratase/isomerase that is required for synthesis of the biofilm dispersion autoinducer cis-2-decenoic acid in the human pathogen Pseudomonas aeruginosa. The protein is encoded by PA14_54640 (PA0745), named dspI for dispersion inducer. The gene sequence for this protein shows significant homology to RpfF in Xanthomonas campestris. Inactivation of dspI was shown to abolish biofilm dispersion autoinduction in continuous cultures of P. aeruginosa and resulted in biofilms that were significantly greater in thickness and biomass than those of the parental wild-type strain. Dispersion was shown to be inducible in dspI mutants by the exogenous addition of synthetic cis-2-decenoic acid or by complementation of ΔdspI in trans under the control of an arabinose-inducible promoter. Mutation of dspI was also shown to abolish cis-2-decenoic acid production, as revealed by gas chromatography-mass spectrometry (GC-MS) analysis of cell-free spent culture medium. The transcript abundance of dspI correlated with cell density, as determined by quantitative reverse transcriptase (RT) PCR. This regulation is consistent with the characterization of cis-2-decenoic acid as a cell-to-cell communication molecule that regulates biofilm dispersion in a cell density-dependent manner.
Figures


References
-
- Davies DG. 1999. Regulation of matrix polymer in biofilm formation and dispersion, p 93–112 In Wingender J, Neu TR, Flemming H-C. (ed), Microbial extrapolymeric substances, characterization, structure and function. Springer-Verlag, Berlin, Germany
-
- Barber CE, Tang JL, Feng JX, Pan MO, Wilson TJG, Slater H, Dow JM, Williams P, Daniels MJ. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24:555–566 - PubMed
-
- Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu J-L, Tay L, Fang RX, Zhang LH. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903–912 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

