In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader
- PMID: 23935074
- PMCID: PMC3794615
- DOI: 10.1093/nar/gkt690
In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader
Abstract
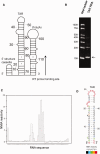
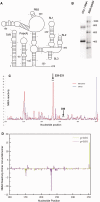
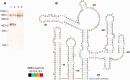
Definitive secondary structural mapping of RNAs in vitro can be complicated by the presence of more than one structural conformer or multimerization of some of the molecules. Until now, probing a single structure of conformationally flexible RNA molecules has typically relied on introducing stabilizing mutations or adjusting buffer conditions or RNA concentration. Here, we present an in-gel SHAPE (selective 2'OH acylation analysed by primer extension) approach, where a mixed structural population of RNA molecules is separated by non-denaturing gel electrophoresis and the conformers are individually probed within the gel matrix. Validation of the technique using a well-characterized RNA stem-loop structure, the HIV-1 trans-activation response element, showed that authentic structure was maintained and that the method was accurate and highly reproducible. To further demonstrate the utility of in-gel SHAPE, we separated and examined monomeric and dimeric species of the HIV-1 packaging signal RNA. Extensive differences in acylation sensitivity were seen between monomer and dimer. The results support a recently proposed structural switch model of RNA genomic dimerization and packaging, and demonstrate the discriminatory power of in-gel SHAPE.
Figures


References
-
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

