The transcription factor Twist1 limits T helper 17 and T follicular helper cell development by repressing the gene encoding the interleukin-6 receptor α chain
- PMID: 23935104
- PMCID: PMC3779737
- DOI: 10.1074/jbc.M113.497248
The transcription factor Twist1 limits T helper 17 and T follicular helper cell development by repressing the gene encoding the interleukin-6 receptor α chain
Abstract
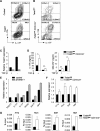
Cytokine responsiveness is a critical component of the ability of cells to respond to the extracellular milieu. Transcription factor-mediated regulation of cytokine receptor expression is a common mode of altering responses to the external environment. We identify the transcription factor Twist1 as a component of a STAT3-induced feedback loop that controls IL-6 signals by directly repressing Il6ra. Human and mouse T cells lacking Twist1 have an increased ability to differentiate into Th17 cells. Mice with a T cell-specific deletion of Twist1 demonstrate increased Th17 and T follicular helper cell development, early onset experimental autoimmune encephalomyelitis, and increased antigen-specific antibody responses. Thus, Twist1 has a critical role in limiting both cell-mediated and humoral immunity.
Keywords: Autoimmunity; Differentiation; Inflammation; Interleukin; STAT Transcription Factor; STAT3; T Cell Biology; T Helper Cell; Transcription Factors.
Figures




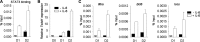

References
-
- Mangan P. R., Harrington L. E., O'Quinn D. B., Helms W. S., Bullard D. C., Elson C. O., Hatton R. D., Wahl S. M., Schoeb T. R., Weaver C. T. (2006) Transforming growth factor-β induces development of the T(H)17 lineage. Nature 441, 231–234 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

