Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability
- PMID: 23935105
- PMCID: PMC3784731
- DOI: 10.1074/jbc.M113.496463
Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability
Abstract
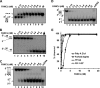
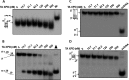
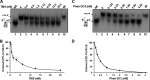
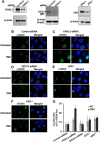
G-quadruplex (G4) DNA, an alternate structure formed by Hoogsteen hydrogen bonds between guanines in G-rich sequences, threatens genomic stability by perturbing normal DNA transactions including replication, repair, and transcription. A variety of G4 topologies (intra- and intermolecular) can form in vitro, but the molecular architecture and cellular factors influencing G4 landscape in vivo are not clear. Helicases that unwind structured DNA molecules are emerging as an important class of G4-resolving enzymes. The BRCA1-associated FANCJ helicase is among those helicases able to unwind G4 DNA in vitro, and FANCJ mutations are associated with breast cancer and linked to Fanconi anemia. FANCJ belongs to a conserved iron-sulfur (Fe S) cluster family of helicases important for genomic stability including XPD (nucleotide excision repair), DDX11 (sister chromatid cohesion), and RTEL (telomere metabolism), genetically linked to xeroderma pigmentosum/Cockayne syndrome, Warsaw breakage syndrome, and dyskeratosis congenita, respectively. To elucidate the role of FANCJ in genomic stability, its molecular functions in G4 metabolism were examined. FANCJ efficiently unwound in a kinetic and ATPase-dependent manner entropically favored unimolecular G4 DNA, whereas other Fe-S helicases tested did not. The G4-specific ligands Phen-DC3 or Phen-DC6 inhibited FANCJ helicase on unimolecular G4 ∼1000-fold better than bi- or tetramolecular G4 DNA. The G4 ligand telomestatin induced DNA damage in human cells deficient in FANCJ but not DDX11 or XPD. These findings suggest FANCJ is a specialized Fe-S cluster helicase that preserves chromosomal stability by unwinding unimolecular G4 DNA likely to form in transiently unwound single-stranded genomic regions.
Keywords: DNA Helicase; DNA Repair; DNA Replication; Fanconi Anemia; G-quadruplex; Genetic Diseases; Genomic Instability.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials
Miscellaneous

