Two-tiered control of epithelial growth and autophagy by the insulin receptor and the ret-like receptor, stitcher
- PMID: 23935447
- PMCID: PMC3720245
- DOI: 10.1371/journal.pbio.1001612
Two-tiered control of epithelial growth and autophagy by the insulin receptor and the ret-like receptor, stitcher
Abstract
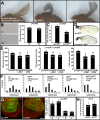
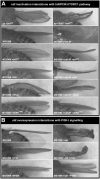
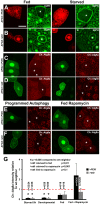
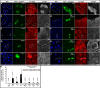
Body size in Drosophila larvae, like in other animals, is controlled by nutrition. Nutrient restriction leads to catabolic responses in the majority of tissues, but the Drosophila mitotic imaginal discs continue growing. The nature of these differential control mechanisms that spare distinct tissues from starvation are poorly understood. Here, we reveal that the Ret-like receptor tyrosine kinase (RTK), Stitcher (Stit), is required for cell growth and proliferation through the PI3K-I/TORC1 pathway in the Drosophila wing disc. Both Stit and insulin receptor (InR) signaling activate PI3K-I and drive cellular proliferation and tissue growth. However, whereas optimal growth requires signaling from both InR and Stit, catabolic changes manifested by autophagy only occur when both signaling pathways are compromised. The combined activities of Stit and InR in ectodermal epithelial tissues provide an RTK-mediated, two-tiered reaction threshold to varying nutritional conditions that promote epithelial organ growth even at low levels of InR signaling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Hietakangas V, Cohen SM (2009) Regulation of tissue growth through nutrient sensing. Annu Rev Genet 43: 389–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

