A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen
- PMID: 23935449
- PMCID: PMC3720256
- DOI: 10.1371/journal.pbio.1001614
A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen
Abstract
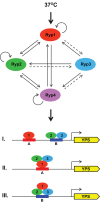
Survival at host temperature is a critical trait for pathogenic microbes of humans. Thermally dimorphic fungal pathogens, including Histoplasma capsulatum, are soil fungi that undergo dramatic changes in cell shape and virulence gene expression in response to host temperature. How these organisms link changes in temperature to both morphologic development and expression of virulence traits is unknown. Here we elucidate a temperature-responsive transcriptional network in H. capsulatum, which switches from a filamentous form in the environment to a pathogenic yeast form at body temperature. The circuit is driven by three highly conserved factors, Ryp1, Ryp2, and Ryp3, that are required for yeast-phase growth at 37°C. Ryp factors belong to distinct families of proteins that control developmental transitions in fungi: Ryp1 is a member of the WOPR family of transcription factors, and Ryp2 and Ryp3 are both members of the Velvet family of proteins whose molecular function is unknown. Here we provide the first evidence that these WOPR and Velvet proteins interact, and that Velvet proteins associate with DNA to drive gene expression. Using genome-wide chromatin immunoprecipitation studies, we determine that Ryp1, Ryp2, and Ryp3 associate with a large common set of genomic loci that includes known virulence genes, indicating that the Ryp factors directly control genes required for pathogenicity in addition to their role in regulating cell morphology. We further dissect the Ryp regulatory circuit by determining that a fourth transcription factor, which we name Ryp4, is required for yeast-phase growth and gene expression, associates with DNA, and displays interdependent regulation with Ryp1, Ryp2, and Ryp3. Finally, we define cis-acting motifs that recruit the Ryp factors to their interwoven network of temperature-responsive target genes. Taken together, our results reveal a positive feedback circuit that directs a broad transcriptional switch between environmental and pathogenic states in response to temperature.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

