Mapping differentiation under mixed culture conditions reveals a tunable continuum of T cell fates
- PMID: 23935451
- PMCID: PMC3728017
- DOI: 10.1371/journal.pbio.1001616
Mapping differentiation under mixed culture conditions reveals a tunable continuum of T cell fates
Abstract
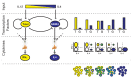
Cell differentiation is typically directed by external signals that drive opposing regulatory pathways. Studying differentiation under polarizing conditions, with only one input signal provided, is limited in its ability to resolve the logic of interactions between opposing pathways. Dissection of this logic can be facilitated by mapping the system's response to mixtures of input signals, which are expected to occur in vivo, where cells are simultaneously exposed to various signals with potentially opposing effects. Here, we systematically map the response of naïve T cells to mixtures of signals driving differentiation into the Th1 and Th2 lineages. We characterize cell state at the single cell level by measuring levels of the two lineage-specific transcription factors (T-bet and GATA3) and two lineage characteristic cytokines (IFN-γ and IL-4) that are driven by these transcription regulators. We find a continuum of mixed phenotypes in which individual cells co-express the two lineage-specific master regulators at levels that gradually depend on levels of the two input signals. Using mathematical modeling we show that such tunable mixed phenotype arises if autoregulatory positive feedback loops in the gene network regulating this process are gradual and dominant over cross-pathway inhibition. We also find that expression of the lineage-specific cytokines follows two independent stochastic processes that are biased by expression levels of the master regulators. Thus, cytokine expression is highly heterogeneous under mixed conditions, with subpopulations of cells expressing only IFN-γ, only IL-4, both cytokines, or neither. The fraction of cells in each of these subpopulations changes gradually with input conditions, reproducing the continuous internal state at the cell population level. These results suggest a differentiation scheme in which cells reflect uncertainty through a continuously tuneable mixed phenotype combined with a biased stochastic decision rather than a binary phenotype with a deterministic decision.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
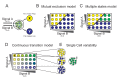
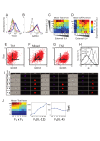

 (see Text S1 for details). In region I, the GRN has a single fixed point with a high level of x and a low level of y. In region II it has a single fixed point with low x and high y. In region III it has a single fixed point with co-expression of both TFs, whereas in region IV it has two stable fixed points (bifurcation). (C–D) TF levels shift continuously upon gradual changes in input signal mixtures. Measured levels (MFI) of T-bet and GATA3 (C) along a trajectory in input plane, which interpolates between a Th1 and a Th2 condition (shown in E, gray line). Continuous changes in TF levels are in agreement with model predictions for n = 1, region III (D) and do not show any bi-stability or sharp transitions as predicted by a high-n model, or low-n model region IV. (E–F) Mapping patterns of TF co-expression over the entire input plane, comparing experiment (E) and model (F). For each TF, we define a threshold level T at ∼50% of its maximal expression level. Regions' color represents patterns of co-expression, as shown in the legend.
(see Text S1 for details). In region I, the GRN has a single fixed point with a high level of x and a low level of y. In region II it has a single fixed point with low x and high y. In region III it has a single fixed point with co-expression of both TFs, whereas in region IV it has two stable fixed points (bifurcation). (C–D) TF levels shift continuously upon gradual changes in input signal mixtures. Measured levels (MFI) of T-bet and GATA3 (C) along a trajectory in input plane, which interpolates between a Th1 and a Th2 condition (shown in E, gray line). Continuous changes in TF levels are in agreement with model predictions for n = 1, region III (D) and do not show any bi-stability or sharp transitions as predicted by a high-n model, or low-n model region IV. (E–F) Mapping patterns of TF co-expression over the entire input plane, comparing experiment (E) and model (F). For each TF, we define a threshold level T at ∼50% of its maximal expression level. Regions' color represents patterns of co-expression, as shown in the legend.

References
-
- Tam PPL, Loebel D a F (2007) Gene function in mouse embryogenesis: get set for gastrulation. Nature Reviews Genetics 8: 368–381 Available at http://www.ncbi.nlm.nih.gov/pubmed/17387317. Accessed 6 March 2013. - PubMed
-
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, et al. (2011) Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240 Available at http://www.ncbi.nlm.nih.gov/pubmed/21295278. Accessed 6 March 2013. - PubMed
-
- Zhou JX, Huang S (2011) Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends in Genetics: TIG 27: 55–62 Available at http://www.ncbi.nlm.nih.gov/pubmed/21146896. Accessed 27 February 2013. - PubMed
-
- Cantor AB, Orkin SH (2001) Hematopoietic development: a balancing act. Current Opinion in Genetics & Development 11: 513–519 Available at http://www.ncbi.nlm.nih.gov/pubmed/11532392. Accessed 6 December 2011. - PubMed
-
- Mariani L, Löhning M, Radbruch A, Höfer T (2004) Transcriptional control networks of cell differentiation: insights from helper T lymphocytes. Progress in Biophysics and Molecular Biology 86: 45–76 Available at http://www.ncbi.nlm.nih.gov/pubmed/15261525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

