Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls
- PMID: 23935489
- PMCID: PMC3723635
- DOI: 10.1371/journal.ppat.1003515
Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls
Abstract
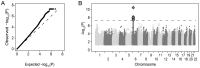
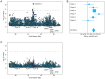
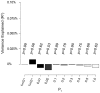
Multiple genome-wide association studies (GWAS) have been performed in HIV-1 infected individuals, identifying common genetic influences on viral control and disease course. Similarly, common genetic correlates of acquisition of HIV-1 after exposure have been interrogated using GWAS, although in generally small samples. Under the auspices of the International Collaboration for the Genomics of HIV, we have combined the genome-wide single nucleotide polymorphism (SNP) data collected by 25 cohorts, studies, or institutions on HIV-1 infected individuals and compared them to carefully matched population-level data sets (a list of all collaborators appears in Note S1 in Text S1). After imputation using the 1,000 Genomes Project reference panel, we tested approximately 8 million common DNA variants (SNPs and indels) for association with HIV-1 acquisition in 6,334 infected patients and 7,247 population samples of European ancestry. Initial association testing identified the SNP rs4418214, the C allele of which is known to tag the HLA-B*57:01 and B*27:05 alleles, as genome-wide significant (p = 3.6 × 10⁻¹¹). However, restricting analysis to individuals with a known date of seroconversion suggested that this association was due to the frailty bias in studies of lethal diseases. Further analyses including testing recessive genetic models, testing for bulk effects of non-genome-wide significant variants, stratifying by sexual or parenteral transmission risk and testing previously reported associations showed no evidence for genetic influence on HIV-1 acquisition (with the exception of CCR5Δ32 homozygosity). Thus, these data suggest that genetic influences on HIV acquisition are either rare or have smaller effects than can be detected by this sample size.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Le Clerc S, Limou S, Coulonges C, Carpentier W, Dina C, et al. (2009) Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03). J Infect Dis 200: 1194–1201. - PubMed
-
- Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, et al. (2009) Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J Infect Dis 199: 419–426. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

