Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets
- PMID: 23935497
- PMCID: PMC3731246
- DOI: 10.1371/journal.ppat.1003513
Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets
Abstract
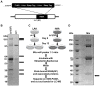
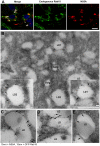
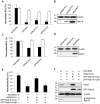
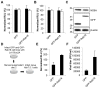
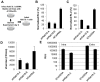
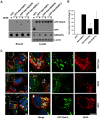
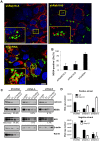
Hepatitis C virus (HCV) is a single-stranded RNA virus that replicates on endoplasmic reticulum-derived membranes. HCV particle assembly is dependent on the association of core protein with cellular lipid droplets (LDs). However, it remains uncertain whether HCV assembly occurs at the LD membrane itself or at closely associated ER membranes. Furthermore, it is not known how the HCV replication complex and progeny genomes physically associate with the presumed sites of virion assembly at or near LDs. Using an unbiased proteomic strategy, we have found that Rab18 interacts with the HCV nonstructural protein NS5A. Rab18 associates with LDs and is believed to promote physical interaction between LDs and ER membranes. Active (GTP-bound) forms of Rab18 bind more strongly to NS5A than a constitutively GDP-bound mutant. NS5A colocalizes with Rab18-positive LDs in HCV-infected cells, and Rab18 appears to promote the physical association of NS5A and other replicase components with LDs. Modulation of Rab18 affects genome replication and possibly also the production of infectious virions. Our results support a model in which specific interactions between viral and cellular proteins may promote the physical interaction between membranous HCV replication foci and lipid droplets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Role for TBC1D20 and Rab1 in hepatitis C virus replication via interaction with lipid droplet-bound nonstructural protein 5A.J Virol. 2012 Jun;86(12):6491-502. doi: 10.1128/JVI.00496-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491470 Free PMC article.
-
Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein.PLoS Pathog. 2013;9(4):e1003302. doi: 10.1371/journal.ppat.1003302. Epub 2013 Apr 11. PLoS Pathog. 2013. PMID: 23593007 Free PMC article.
-
Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication.J Virol. 2014 Jun;88(12):6793-804. doi: 10.1128/JVI.00045-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696471 Free PMC article.
-
Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex.Antiviral Res. 2003 Oct;60(2):103-9. doi: 10.1016/j.antiviral.2003.08.017. Antiviral Res. 2003. PMID: 14638405 Review.
-
Hepatitis C virus: viral proteins on the move.Biochem Soc Trans. 2009 Oct;37(Pt 5):986-90. doi: 10.1042/BST0370986. Biochem Soc Trans. 2009. PMID: 19754437 Review.
Cited by
-
The use of novel epitope-tagged arenaviruses reveals that Rab5c-positive endosomal membranes are targeted by the LCMV matrix protein.J Gen Virol. 2018 Feb;99(2):187-193. doi: 10.1099/jgv.0.001004. J Gen Virol. 2018. PMID: 29393022 Free PMC article.
-
Hepatitis C Virus NS5A Activates Mitophagy Through Cargo Receptor and Phagophore Formation.Pathogens. 2024 Dec 23;13(12):1139. doi: 10.3390/pathogens13121139. Pathogens. 2024. PMID: 39770398 Free PMC article.
-
Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion.J Virol. 2016 Jul 27;90(16):7159-70. doi: 10.1128/JVI.00826-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27226379 Free PMC article.
-
Hepatitis C virus promotes virion secretion through cleavage of the Rab7 adaptor protein RILP.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):12484-12489. doi: 10.1073/pnas.1607277113. Epub 2016 Oct 17. Proc Natl Acad Sci U S A. 2016. PMID: 27791088 Free PMC article.
-
Forkhead box transcription factor regulation and lipid accumulation by hepatitis C virus.J Virol. 2014 Apr;88(8):4195-203. doi: 10.1128/JVI.03327-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478438 Free PMC article.
References
-
- Brass V, Bieck E, Montserret R, Wölk B, Hellings JA, et al. (2002) An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem 277: 8130–8139. - PubMed
-
- Penin F, Brass V, Appel N, Ramboarina S, Montserret R, et al. (2004) Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem 279: 40835–40843. - PubMed
-
- Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM (2004) The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem 279: 48576–48587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

