Transposon domestication versus mutualism in ciliate genome rearrangements
- PMID: 23935529
- PMCID: PMC3731211
- DOI: 10.1371/journal.pgen.1003659
Transposon domestication versus mutualism in ciliate genome rearrangements
Abstract
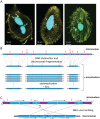
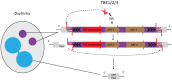
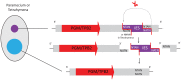
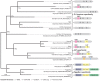
Ciliated protists rearrange their genomes dramatically during nuclear development via chromosome fragmentation and DNA deletion to produce a trimmer and highly reorganized somatic genome. The deleted portion of the genome includes potentially active transposons or transposon-like sequences that reside in the germline. Three independent studies recently showed that transposase proteins of the DDE/DDD superfamily are indispensible for DNA processing in three distantly related ciliates. In the spirotrich Oxytricha trifallax, high copy-number germline-limited transposons mediate their own excision from the somatic genome but also contribute to programmed genome rearrangement through a remarkable transposon mutualism with the host. By contrast, the genomes of two oligohymenophorean ciliates, Tetrahymena thermophila and Paramecium tetraurelia, encode homologous PiggyBac-like transposases as single-copy genes in both their germline and somatic genomes. These domesticated transposases are essential for deletion of thousands of different internal sequences in these species. This review contrasts the events underlying somatic genome reduction in three different ciliates and considers their evolutionary origins and the relationships among their distinct mechanisms for genome remodeling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Orgel LE, Crick FH (1980) Selfish DNA: the ultimate parasite. Nature 284: 604–607. - PubMed
-
- Kazazian HH Jr (2004) Mobile elements: drivers of genome evolution. Science 303: 1626–1632. - PubMed
-
- Volff JN (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28: 913–922. - PubMed
-
- Kapitonov VV, Jurka J (2005) RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 3: e181 doi:10.1371/journal.pbio.0030181 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

