Assumptions and properties of limiting pathway models for analysis of epistasis in complex traits
- PMID: 23935903
- PMCID: PMC3728313
- DOI: 10.1371/journal.pone.0068913
Assumptions and properties of limiting pathway models for analysis of epistasis in complex traits
Abstract
For most complex traits, results from genome-wide association studies show that the proportion of the phenotypic variance attributable to the additive effects of individual SNPs, that is, the heritability explained by the SNPs, is substantially less than the estimate of heritability obtained by standard methods using correlations between relatives. This difference has been called the "missing heritability". One explanation is that heritability estimates from family (including twin) studies are biased upwards. Zuk et al. revisited overestimation of narrow sense heritability from twin studies as a result of confounding with non-additive genetic variance. They propose a limiting pathway (LP) model that generates significant epistatic variation and its simple parametrization provides a convenient way to explore implications of epistasis. They conclude that over-estimation of narrow sense heritability from family data ('phantom heritability') may explain an important proportion of missing heritability. We show that for highly heritable quantitative traits large phantom heritability estimates from twin studies are possible only if a large contribution of common environment is assumed. The LP model is underpinned by strong assumptions that are unlikely to hold, including that all contributing pathways have the same mean and variance and are uncorrelated. Here, we relax the assumptions that underlie the LP model to be more biologically plausible. Together with theoretical, empirical, and pragmatic arguments we conclude that in outbred populations the contribution of additive genetic variance is likely to be much more important than the contribution of non-additive variance.
Conflict of interest statement
Figures

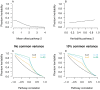
 , contribution of common family environment is zero (i.e.,
, contribution of common family environment is zero (i.e.,  ), pathway heritability
), pathway heritability  , pathway correlations
, pathway correlations  .
.References
-
- Maher B (2008) The case of the missing heritability. Nature 456: 18–21. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

