Structure of the SCAN domain of human paternally expressed gene 3 protein
- PMID: 23936039
- PMCID: PMC3720700
- DOI: 10.1371/journal.pone.0069538
Structure of the SCAN domain of human paternally expressed gene 3 protein
Abstract
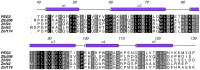
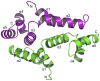
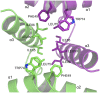
Human paternally expressed gene 3 protein (PEG3) is a large multi-domain entity with diverse biological functions, including acting as a transcription factor. PEG3 contains twelve Cys2-His2 type zinc finger domains, extended regions of predicted disorder and at the N-terminus a SCAN domain. PEG3 has been identified as partner of the E3 ubiquitin-protein ligase Siah1, an association we sought to investigate. An efficient bacterial recombinant expression system of the human PEG3-SCAN domain was prepared and crystals appeared spontaneously when the protein was being concentrated after purification. The structure was determined at 1.95 Å resolution and reveals a polypeptide fold of five helices in an extended configuration. An extensive dimerization interface, using almost a quarter of the solvent accessible surface, and key salt bridge interactions explain the stability of the dimer. Comparison with other SCAN domains reveals a high degree of conservation involving residues that contribute to the dimer interface. The PEG3-SCAN domain appears to constitute an assembly block, enabling PEG3 homo- or heterodimerization to control gene expression in a combinatorial fashion.
Conflict of interest statement
Figures





References
-
- Relaix F, Weng X, Marazzi G, Yang E, Copeland N, et al. (1996) Pw1, a novel zinc finger gene implicated in the myogenic and neuronal lineages. Dev Biol 177: 383–396. - PubMed
-
- Lee MS, Gippert GP, Soman KV, Case DA, Wright PE (1989) Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 245: 635–637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

