Cep63 and cep152 cooperate to ensure centriole duplication
- PMID: 23936128
- PMCID: PMC3728344
- DOI: 10.1371/journal.pone.0069986
Cep63 and cep152 cooperate to ensure centriole duplication
Abstract
Centrosomes consist of two centrioles embedded in pericentriolar material and function as the main microtubule organising centres in dividing animal cells. They ensure proper formation and orientation of the mitotic spindle and are therefore essential for the maintenance of genome stability. Centrosome function is crucial during embryonic development, highlighted by the discovery of mutations in genes encoding centrosome or spindle pole proteins that cause autosomal recessive primary microcephaly, including Cep63 and Cep152. In this study we show that Cep63 functions to ensure that centriole duplication occurs reliably in dividing mammalian cells. We show that the interaction between Cep63 and Cep152 can occur independently of centrosome localisation and that the two proteins are dependent on one another for centrosomal localisation. Further, both mouse and human Cep63 and Cep152 cooperate to ensure efficient centriole duplication by promoting the accumulation of essential centriole duplication factors upstream of SAS-6 recruitment and procentriole formation. These observations describe the requirement for Cep63 in maintaining centriole number in dividing mammalian cells and further establish the order of events in centriole formation.
Conflict of interest statement
Figures
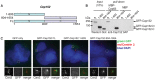

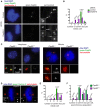
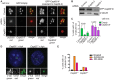

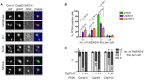
References
-
- Gönczy P (2012) Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol 13: 425–435. - PubMed
-
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678. - PubMed
-
- Nigg EA (2007) Centrosome duplication: of rules and licenses. Trends Cell Biol 17: 215–221. - PubMed
-
- Strnad P, Gönczy P (2008) Mechanisms of procentriole formation. Trends Cell Biol 18: 389–396. - PubMed
-
- Delattre M, Canard C, Gönczy P (2006) Sequential protein recruitment in C. elegans centriole formation. Curr Biol 16: 1844–1849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

