Conformational stability of fibrillar amyloid-beta oligomers via protofilament pair formation - a systematic computational study
- PMID: 23936224
- PMCID: PMC3729696
- DOI: 10.1371/journal.pone.0070521
Conformational stability of fibrillar amyloid-beta oligomers via protofilament pair formation - a systematic computational study
Abstract
Amyloid-[Formula: see text] (A[Formula: see text]) oligomers play a crucial role in Alzheimer's disease due to their neurotoxic aggregation properties. Fibrillar A[Formula: see text] oligomerization can lead to protofilaments and protofilament pairs via oligomer elongation and oligomer association, respectively. Small fibrillar oligomers adopt the protofilament topology, whereas fibrils contain at least protofilament pairs. To date, the underlying growth mechanism from oligomers to the mature fibril still remains to be elucidated. Here, we performed all-atom molecular dynamics simulations in explicit solvent on single layer-like protofilaments and fibril-like protofilament pairs of different size ranging from the tetramer to the 48-mer. We found that the initial U-shaped topology per monomer is maintained over time in all oligomers. The observed deviations of protofilaments from the starting structure increase significantly with size due to the twisting of the in-register parallel [Formula: see text]-sheets. This twist causes long protofilaments to be unstable and leads to a breakage. Protofilament pairs, which are stabilized by a hydrophobic interface, exhibit more fibril-like properties such as the overall structure and the twist angle. Thus, they can act as stable conformational templates for further fibril growth. Key properties like the twist angle, shape complementarity, and energetics show a size-dependent behavior so that small oligomers favor the protofilament topology, whereas large oligomers favor the protofilament pair topology. The region for this conformational transition is at the size of approximately twelve A[Formula: see text] monomers. From that, we propose the following growth mechanism from A[Formula: see text] oligomers to fibrils: (1) elongation of short protofilaments; (2) breakage of large protofilaments; (3) formation of short protofilament pairs; and (4) elongation of protofilament pairs.
Conflict of interest statement
Figures

 ) as an example for the orientation of peptide chains within the protofilaments. (C) The interaction between hydrophobic residues around M35 (yellow) in the C-termini of two opposite protofilaments constitutes the interface in the protofilament pairs. (D) The 8-mer (O
) as an example for the orientation of peptide chains within the protofilaments. (C) The interaction between hydrophobic residues around M35 (yellow) in the C-termini of two opposite protofilaments constitutes the interface in the protofilament pairs. (D) The 8-mer (O ) as an example for the orientation of peptide chains in the protofilament pairs. (E) Two different angles were analyzed, the twist angle and the angle between adjacent monomers. (F) Two oligomers can either be combined to form a longer protofilament (elongation) or be merged via C-terminal contacts to form a protofilament pair (thickening). Therefore, two types of MM/GBSA calculations were performed: segmentation of protofilaments along the red plane and segmentation of protofilament pairs along the blue plane.
) as an example for the orientation of peptide chains in the protofilament pairs. (E) Two different angles were analyzed, the twist angle and the angle between adjacent monomers. (F) Two oligomers can either be combined to form a longer protofilament (elongation) or be merged via C-terminal contacts to form a protofilament pair (thickening). Therefore, two types of MM/GBSA calculations were performed: segmentation of protofilaments along the red plane and segmentation of protofilament pairs along the blue plane.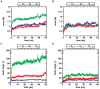
 oligomers. Upon formation of the C-terminal interface leading to protofilament pairs (B), the rmsd shows no difference between the small, medium or large system. Parallel in-register
oligomers. Upon formation of the C-terminal interface leading to protofilament pairs (B), the rmsd shows no difference between the small, medium or large system. Parallel in-register  -sheets reveal a general twist along the growth axis. The twist angle increases with size in the protofilaments (C) and is the reason for the rather high rmsd values (A). Upon formation of the C-terminal interface leading to protofilament pairs (D), the twist angle remains stable over time, indicating that addition of a second layer counteracts twisting of parallel
-sheets reveal a general twist along the growth axis. The twist angle increases with size in the protofilaments (C) and is the reason for the rather high rmsd values (A). Upon formation of the C-terminal interface leading to protofilament pairs (D), the twist angle remains stable over time, indicating that addition of a second layer counteracts twisting of parallel  -sheets.
-sheets.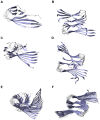
 , but the hydrophobic residues in the C-terminus are covered by the second layer. (C) The protofilament hexamer (O
, but the hydrophobic residues in the C-terminus are covered by the second layer. (C) The protofilament hexamer (O ) displays the large twist of the parallel
) displays the large twist of the parallel  -sheets. (D) The protofilament pair dodecamer (O
-sheets. (D) The protofilament pair dodecamer (O ) has a smaller twist angle than the protofilament hexamer due to the conteracting stabilization by the C-terminal interaction. (E) The protofilament 24-mer (O
) has a smaller twist angle than the protofilament hexamer due to the conteracting stabilization by the C-terminal interaction. (E) The protofilament 24-mer (O ) shows a small angle between adjacent monomers but the large overall twist angle. (F) The protofilament pair 48-mer (O
) shows a small angle between adjacent monomers but the large overall twist angle. (F) The protofilament pair 48-mer (O ) shows that the overall twist angle is reduced upon C-terminal interaction.
) shows that the overall twist angle is reduced upon C-terminal interaction.
 -sheets. N-terminal and C-terminal
-sheets. N-terminal and C-terminal  -sheet of each monomer are separated by a turn region. See Figures S13–S22 in File SI for the results of all other oligomers.
-sheet of each monomer are separated by a turn region. See Figures S13–S22 in File SI for the results of all other oligomers.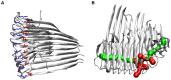
 ), the side chains of D23 and K28 are depicted as red and blue sticks, respectively. (B) Visualization of the water channel in the protofilament 12-mer (O
), the side chains of D23 and K28 are depicted as red and blue sticks, respectively. (B) Visualization of the water channel in the protofilament 12-mer (O , 50 ns); main entrance channel (green) and channel exits through neighboring turns (red).
, 50 ns); main entrance channel (green) and channel exits through neighboring turns (red).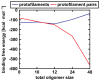

 -sheets initiates a breakage into oligomers of smaller size. Breakage points in the 24-mer (A) and the 48-mer (B) are indicated with red arrows.
-sheets initiates a breakage into oligomers of smaller size. Breakage points in the 24-mer (A) and the 48-mer (B) are indicated with red arrows.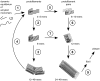
 via elongation of large protofilament pairs to fibrils and plaques.
via elongation of large protofilament pairs to fibrils and plaques.References
-
- Alzheimer A (1907) Über eine eigenartige Erkrankung der Hirnrinde. Allg Zschr Psychiat 64: 146–148.
-
- Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297: 353–356. - PubMed
-
- Selkoe DJ (2004) Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol 6: 1054–1061. - PubMed
-
- Haass C, Selkoe DJ (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol 8: 101–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

