HIV-1 tropism testing in subjects achieving undetectable HIV-1 RNA: diagnostic accuracy, viral evolution and compartmentalization
- PMID: 23936293
- PMCID: PMC3731261
- DOI: 10.1371/journal.pone.0067085
HIV-1 tropism testing in subjects achieving undetectable HIV-1 RNA: diagnostic accuracy, viral evolution and compartmentalization
Abstract
Background: Technically, HIV-1 tropism can be evaluated in plasma or peripheral blood mononuclear cells (PBMCs). However, only tropism testing of plasma HIV-1 has been validated as a tool to predict virological response to CCR5 antagonists in clinical trials. The preferable tropism testing strategy in subjects with undetectable HIV-1 viremia, in whom plasma tropism testing is not feasible, remains uncertain.
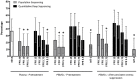
Methods & results: We designed a proof-of-concept study including 30 chronically HIV-1-infected individuals who achieved HIV-1 RNA <50 copies/mL during at least 2 years after first-line ART initiation. First, we determined the diagnostic accuracy of 454 and population sequencing of gp120 V3-loops in plasma and PBMCs, as well as of MT-2 assays before ART initiation. The Enhanced Sensitivity Trofile Assay (ESTA) was used as the technical reference standard. 454 sequencing of plasma viruses provided the highest agreement with ESTA. The accuracy of 454 sequencing decreased in PBMCs due to reduced specificity. Population sequencing in plasma and PBMCs was slightly less accurate than plasma 454 sequencing, being less sensitive but more specific. MT-2 assays had low sensitivity but 100% specificity. Then, we used optimized 454 sequence data to investigate viral evolution in PBMCs during viremia suppression and only found evolution of R5 viruses in one subject. No de novo CXCR4-using HIV-1 production was observed over time. Finally, Slatkin-Maddison tests suggested that plasma and cell-associated V3 forms were sometimes compartmentalized.
Conclusions: The absence of tropism shifts during viremia suppression suggests that, when available, testing of stored plasma samples is generally safe and informative, provided that HIV-1 suppression is maintained. Tropism testing in PBMCs may not necessarily produce equivalent biological results to plasma, because the structure of viral populations and the diagnostic performance of tropism assays may sometimes vary between compartments. Thereby, proviral DNA tropism testing should be specifically validated in clinical trials before it can be applied to routine clinical decision-making.
Conflict of interest statement
Figures


References
-
- Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, et al. (2006) Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol 80: 4909–4920. - PMC - PubMed
-
- Cooper DA, Heera J, Goodrich J, Tawadrous M, Saag M, et al. (2010) Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J Infect Dis 201: 803–813. - PubMed
-
- Fatkenheuer G, Nelson M, Lazzarin A, Konourina I, Hoepelman AI, et al. (2008) Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med 359: 1442–1455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

