A possible mechanism behind autoimmune disorders discovered by genome-wide linkage and association analysis in celiac disease
- PMID: 23936387
- PMCID: PMC3732286
- DOI: 10.1371/journal.pone.0070174
A possible mechanism behind autoimmune disorders discovered by genome-wide linkage and association analysis in celiac disease
Abstract
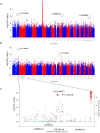
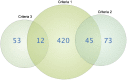
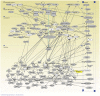
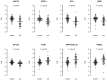
Celiac disease is a common autoimmune disorder characterized by an intestinal inflammation triggered by gluten, a storage protein found in wheat, rye and barley. Similar to other autoimmune diseases such as type 1 diabetes, psoriasis and rheumatoid arthritis, celiac disease is the result of an immune response to self-antigens leading to tissue destruction and production of autoantibodies. Common diseases like celiac disease have a complex pattern of inheritance with inputs from both environmental as well as additive and non-additive genetic factors. In the past few years, Genome Wide Association Studies (GWAS) have been successful in finding genetic risk variants behind many common diseases and traits. To complement and add to the previous findings, we performed a GWAS including 206 trios from 97 nuclear Swedish and Norwegian families affected with celiac disease. By stratifying for HLA-DQ, we identified a new genome-wide significant risk locus covering the DUSP10 gene. To further investigate the associations from the GWAS we performed pathway analyses and two-locus interaction analyses. These analyses showed an over-representation of genes involved in type 2 diabetes and identified a set of candidate mechanisms and genes of which some were selected for mRNA expression analysis using small intestinal biopsies from 98 patients. Several genes were expressed differently in the small intestinal mucosa from patients with celiac autoimmunity compared to intestinal mucosa from control patients. From top-scoring regions we identified susceptibility genes in several categories: 1) polarity and epithelial cell functionality; 2) intestinal smooth muscle; 3) growth and energy homeostasis, including proline and glutamine metabolism; and finally 4) innate and adaptive immune system. These genes and pathways, including specific functions of DUSP10, together reveal a new potential biological mechanism that could influence the genesis of celiac disease, and possibly also other chronic disorders with an inflammatory component.
Conflict of interest statement
Figures


References
-
- Myleus A, Ivarsson A, Webb C, Danielsson L, Hernell O, et al. (2009) Celiac disease revealed in 3% of Swedish 12-year-olds born during an epidemic. J Pediatr Gastroenterol Nutr 49: 170–176. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

