Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1β and tumor necrosis factor α
- PMID: 23936458
- PMCID: PMC3731334
- DOI: 10.1371/journal.pone.0070622
Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1β and tumor necrosis factor α
Abstract
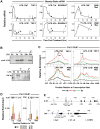
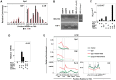
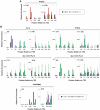
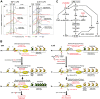
Interleukin-1β and Tumor Necrosis Factor α play related, but distinct, roles in immunity and disease. Our study revealed major mechanistic distinctions in the Toll-like receptor (TLR) signaling-dependent induction for the rapidly expressed genes (IL1B and TNF) coding for these two cytokines. Prior to induction, TNF exhibited pre-bound TATA Binding Protein (TBP) and paused RNA Polymerase II (Pol II), hallmarks of poised immediate-early (IE) genes. In contrast, unstimulated IL1B displayed very low levels of both TBP and paused Pol II, requiring the lineage-specific Spi-1/PU.1 (Spi1) transcription factor as an anchor for induction-dependent interaction with two TLR-activated transcription factors, C/EBPβ and NF-κB. Activation and DNA binding of these two pre-expressed factors resulted in de novo recruitment of TBP and Pol II to IL1B in concert with a permissive state for elongation mediated by the recruitment of elongation factor P-TEFb. This Spi1-dependent mechanism for IL1B transcription, which is unique for a rapidly-induced/poised IE gene, was more dependent upon P-TEFb than was the case for the TNF gene. Furthermore, the dependence on phosphoinositide 3-kinase for P-TEFb recruitment to IL1B paralleled a greater sensitivity to the metabolic state of the cell and a lower sensitivity to the phenomenon of endotoxin tolerance than was evident for TNF. Such differences in induction mechanisms argue against the prevailing paradigm that all IE genes possess paused Pol II and may further delineate the specific roles played by each of these rapidly expressed immune modulators.
Conflict of interest statement
Figures




References
-
- Foster SL, Hargreaves DC, Medzhitov R (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447: 972–978. - PubMed
-
- Fenton MJ, Vermeulen MW, Clark BD, Webb AC, Auron PE (1988) Human pro-IL-1b gene expression in monocytic cells is regulated by two distinct pathways. J Immunol 140: 2267–2273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

