Redox signaling mediated by the gut microbiota
- PMID: 23937589
- PMCID: PMC5131718
- DOI: 10.3109/10715762.2013.833331
Redox signaling mediated by the gut microbiota
Abstract
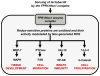
The microbiota that occupies the mammalian intestine can modulate a range of physiological functions, including control over immune responses, epithelial barrier function, and cellular proliferation. While commensal prokaryotic organisms are well known to stimulate inflammatory signaling networks, less is known about control over homeostatic pathways. Recent work has shown that gut epithelia contacted by enteric commensal bacteria rapidly generate reactive oxygen species (ROS). While the induced production of ROS in professional phagocytes via stimulation of formyl peptide receptors (FPRs) and activation of NADPH oxidase 2 (Nox2) is a well-studied process, ROS are also similarly elicited in other cell types, including intestinal epithelia, in response to microbial signals via FPRs and the epithelial NADPH oxidase 1 (Nox1). ROS generated by Nox enzymes have been shown to function as critical second messengers in multiple signal transduction pathways via the rapid and transient oxidative inactivation of a distinct class of sensor proteins bearing oxidant-sensitive thiol groups. These redox-sensitive proteins include tyrosine phosphatases that serve as regulators of MAP kinase pathways, focal adhesion kinase, as well as components involved in NF-κB activation. As microbe-elicited ROS has been shown to stimulate cellular proliferation and motility, and to modulate innate immune signaling, we hypothesize that many of the established effects of the normal microbiota on intestinal physiology may be at least partially mediated by this ROS-dependent mechanism.
Figures

References
-
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5(7):e156. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
