Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA
- PMID: 23937650
- PMCID: PMC3751371
- DOI: 10.1186/1471-2164-14-543
Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA
Abstract
Background: Epstein-Barr virus (EBV) is a human herpesvirus implicated in cancer and autoimmune disorders. Little is known concerning the roles of RNA structure in this important human pathogen. This study provides the first comprehensive genome-wide survey of RNA and RNA structure in EBV.
Results: Novel EBV RNAs and RNA structures were identified by computational modeling and RNA-Seq analyses of EBV. Scans of the genomic sequences of four EBV strains (EBV-1, EBV-2, GD1, and GD2) and of the closely related Macacine herpesvirus 4 using the RNAz program discovered 265 regions with high probability of forming conserved RNA structures. Secondary structure models are proposed for these regions based on a combination of free energy minimization and comparative sequence analysis. The analysis of RNA-Seq data uncovered the first observation of a stable intronic sequence RNA (sisRNA) in EBV. The abundance of this sisRNA rivals that of the well-known and highly expressed EBV-encoded non-coding RNAs (EBERs).
Conclusion: This work identifies regions of the EBV genome likely to generate functional RNAs and RNA structures, provides structural models for these regions, and discusses potential functions suggested by the modeled structures. Enhanced understanding of the EBV transcriptome will guide future experimental analyses of the discovered RNAs and RNA structures.
Figures

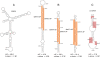





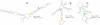
References
-
- Epstein-Barr virus and infectious mononucleosis. http://www.cdc.gov/ncidod/diseases/ebv.htm.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

