Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations
- PMID: 23937694
- PMCID: PMC3935411
- DOI: 10.1111/tpj.12307
Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations
Abstract
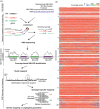
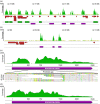
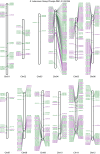
RenSeq is a NB-LRR (nucleotide binding-site leucine-rich repeat) gene-targeted, Resistance gene enrichment and sequencing method that enables discovery and annotation of pathogen resistance gene family members in plant genome sequences. We successfully applied RenSeq to the sequenced potato Solanum tuberosum clone DM, and increased the number of identified NB-LRRs from 438 to 755. The majority of these identified R gene loci reside in poorly or previously unannotated regions of the genome. Sequence and positional details on the 12 chromosomes have been established for 704 NB-LRRs and can be accessed through a genome browser that we provide. We compared these NB-LRR genes and the corresponding oligonucleotide baits with the highest sequence similarity and demonstrated that ~80% sequence identity is sufficient for enrichment. Analysis of the sequenced tomato S. lycopersicum 'Heinz 1706' extended the NB-LRR complement to 394 loci. We further describe a methodology that applies RenSeq to rapidly identify molecular markers that co-segregate with a pathogen resistance trait of interest. In two independent segregating populations involving the wild Solanum species S. berthaultii (Rpi-ber2) and S. ruiz-ceballosii (Rpi-rzc1), we were able to apply RenSeq successfully to identify markers that co-segregate with resistance towards the late blight pathogen Phytophthora infestans. These SNP identification workflows were designed as easy-to-adapt Galaxy pipelines.
Keywords: NB-LRR; Solanaceae; Solanum berthaultii; Solanum lycopersicum; Solanum ruiz-ceballosii; Solanum tuberosum Group Phureja clone DM1-3 516 R44; next-generation sequencing; pathogen resistance; target enrichment; technical advance.
© 2013 The Authors The Plant Journal © 2013 John Wiley & Sons Ltd.
Figures


References
-
- Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4(11):903–905. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. - PubMed
-
- Andolfo G, Sanseverino W, Rombauts S, Van de Peer Y, Bradeen JM, Carputo D, Frusciante L, Ercolano MR. Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol. 2013;197(1):223–237. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/C007522/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- SCR/929/11/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G02197X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H019820/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

