The Vertebrate Trait Ontology: a controlled vocabulary for the annotation of trait data across species
- PMID: 23937709
- PMCID: PMC3851175
- DOI: 10.1186/2041-1480-4-13
The Vertebrate Trait Ontology: a controlled vocabulary for the annotation of trait data across species
Abstract
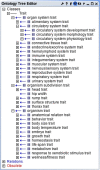
Background: The use of ontologies to standardize biological data and facilitate comparisons among datasets has steadily grown as the complexity and amount of available data have increased. Despite the numerous ontologies available, one area currently lacking a robust ontology is the description of vertebrate traits. A trait is defined as any measurable or observable characteristic pertaining to an organism or any of its substructures. While there are several ontologies to describe entities and processes in phenotypes, diseases, and clinical measurements, one has not been developed for vertebrate traits; the Vertebrate Trait Ontology (VT) was created to fill this void.
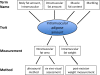
Description: Significant inconsistencies in trait nomenclature exist in the literature, and additional difficulties arise when trait data are compared across species. The VT is a unified trait vocabulary created to aid in the transfer of data within and between species and to facilitate investigation of the genetic basis of traits. Trait information provides a valuable link between the measurements that are used to assess the trait, the phenotypes related to the traits, and the diseases associated with one or more phenotypes. Because multiple clinical and morphological measurements are often used to assess a single trait, and a single measurement can be used to assess multiple physiological processes, providing investigators with standardized annotations for trait data will allow them to investigate connections among these data types.
Conclusions: The annotation of genomic data with ontology terms provides unique opportunities for data mining and analysis. Links between data in disparate databases can be identified and explored, a strategy that is particularly useful for cross-species comparisons or in situations involving inconsistent terminology. The VT provides a common basis for the description of traits in multiple vertebrate species. It is being used in the Rat Genome Database and Animal QTL Database for annotation of QTL data for rat, cattle, chicken, swine, sheep, and rainbow trout, and in the Mouse Phenome Database to annotate strain characterization data. In these databases, data are also cross-referenced to applicable terms from other ontologies, providing additional avenues for data mining and analysis. The ontology is available at http://bioportal.bioontology.org/ontologies/50138.
Figures

References
-
- Haendel M, Neuhaus F, Osumi-Sutherland D, Mabee P, Mejino JJ, Mungall C, Smith B. In: Anatomy Ontologies for Bioinformatics: Principles and Practice. Burger A, Davidson D, Baldock R, editor. London: Springer; 2008. CARO - The Common Anatomy Reference Ontology; pp. 327–350.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

