Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition
- PMID: 23938295
- PMCID: PMC3817542
- DOI: 10.1038/cr.2013.110
Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition
Abstract
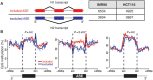
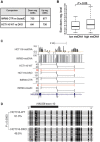
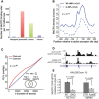
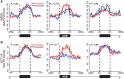
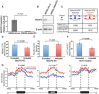
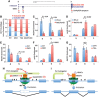
Although the function of DNA methylation in gene promoter regions is well established in transcriptional repression, the function of the evolutionarily conserved widespread distribution of DNA methylation in gene body regions remains incompletely understood. Here, we show that DNA methylation is enriched in included alternatively spliced exons (ASEs), and that inhibition of DNA methylation results in aberrant splicing of ASEs. The methyl-CpG-binding protein MeCP2 is enriched in included ASEs, particularly those that are also highly methylated, and inhibition of DNA methylation disrupts specific targeting of MeCP2 to exons. Interestingly, ablation of MeCP2 results in increased histone acetylation and aberrant ASE-skipping events. We further show that inhibition of histone deacetylase (HDAC) activity leads to exon skipping that shows a highly significant degree of overlap with that caused by MeCP2 knockdown. Together, our data indicate that intragenic DNA methylation operates in exon definition to modulate alternative RNA splicing and can enhance exon recognition via recruitment of the multifunctional protein MeCP2, which thereby maintains local histone hypoacetylation through the subsequent recruitment of HDACs.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

