'There and back again': revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era
- PMID: 23938753
- PMCID: PMC3758188
- DOI: 10.1098/rstb.2012.0504
'There and back again': revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era
Abstract
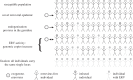
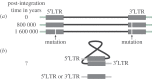
Almost 8% of the human genome comprises endogenous retroviruses (ERVs). While they have been shown to cause specific pathologies in animals, such as cancer, their association with disease in humans remains controversial. The limited evidence is partly due to the physical and bioethical restrictions surrounding the study of transposons in humans, coupled with the major experimental and bioinformatics challenges surrounding the association of ERVs with disease in general. Two biotechnological landmarks of the past decade provide us with unprecedented research artillery: (i) the ultra-fine sequencing of the human genome and (ii) the emergence of high-throughput sequencing technologies. Here, we critically assemble research about potential pathologies of ERVs in humans. We argue that the time is right to revisit the long-standing questions of human ERV pathogenesis within a robust and carefully structured framework that makes full use of genomic sequence data. We also pose two thought-provoking research questions on potential pathophysiological roles of ERVs with respect to immune escape and regulation.
Keywords: HERV-K; endogenous retroviruses; pathogenesis; pathophysiology.
Figures

References
-
- Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42, 709–732 (doi:10.1146/annurev.genet.42.110807.091501) - DOI - PubMed
-
- Boeke JD, Stoye JP. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In Retroviruses (eds Coffin JM, Hughes SH, Varmus HE.), pp. 343–436 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press - PubMed
-
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (doi:10.1038/35057062) - DOI - PubMed
-
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (doi:10.1038/nature11632) - DOI - PMC - PubMed
-
- Ioannidis JP. 2005. Why most published research findings are false. PLoS Med. 2, e124 (doi:10.1371/journal.pmed.0020124) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
