Evolutionary analysis of hepatitis C virus gene sequences from 1953
- PMID: 23938759
- PMCID: PMC3758194
- DOI: 10.1098/rstb.2013.0168
Evolutionary analysis of hepatitis C virus gene sequences from 1953
Abstract
Reconstructing the transmission history of infectious diseases in the absence of medical or epidemiological records often relies on the evolutionary analysis of pathogen genetic sequences. The precision of evolutionary estimates of epidemic history can be increased by the inclusion of sequences derived from 'archived' samples that are genetically distinct from contemporary strains. Historical sequences are especially valuable for viral pathogens that circulated for many years before being formally identified, including HIV and the hepatitis C virus (HCV). However, surprisingly few HCV isolates sampled before discovery of the virus in 1989 are currently available. Here, we report and analyse two HCV subgenomic sequences obtained from infected individuals in 1953, which represent the oldest genetic evidence of HCV infection. The pairwise genetic diversity between the two sequences indicates a substantial period of HCV transmission prior to the 1950s, and their inclusion in evolutionary analyses provides new estimates of the common ancestor of HCV in the USA. To explore and validate the evolutionary information provided by these sequences, we used a new phylogenetic molecular clock method to estimate the date of sampling of the archived strains, plus the dates of four more contemporary reference genomes. Despite the short fragments available, we conclude that the archived sequences are consistent with a proposed sampling date of 1953, although statistical uncertainty is large. Our cross-validation analyses suggest that the bias and low statistical power observed here likely arise from a combination of high evolutionary rate heterogeneity and an unstructured, star-like phylogeny. We expect that attempts to date other historical viruses under similar circumstances will meet similar problems.
Keywords: molecular epidemiology; phylogenetics.
Figures
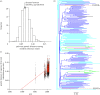
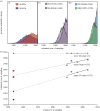
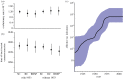
References
-
- Pybus OG, Rambaut A. 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 10, 540–550 (doi:10.1038/nrg2583) - DOI - PMC - PubMed
-
- Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. 2003. Measurably evolving populations. Trends Ecol. Evol. 18, 481–488 (doi:10.1016/S0169-5347(03)00216-7) - DOI
-
- Taubenberger JK, Reid AH, Krafft AE, Bijwaard KE, Fanning TG. 1997. Initial genetic characterization of the 1918 ‘Spanish’ influenza virus. Science 275, 1793–1796 (doi:10.1126/science.275.5307.1793) - DOI - PubMed
-
- Biagini P, et al. 2012. Variola virus in a 300-year-old Siberian mummy. N. Engl. J. Med. 367, 2057–2059 (doi:10.1056/NEJMc1208124) - DOI - PubMed
-
- Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6, e1001191 (doi:10.1371/journal.pgen.1001191) - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

