Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29
- PMID: 23939028
- PMCID: PMC5871871
- DOI: 10.4161/auto.25687
Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29
Abstract
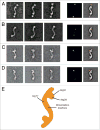
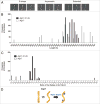
Atg17, in complex with Atg29 and Atg31, constitutes a key module of the Atg1 kinase signaling complex and functions as an important organizer of the phagophore assembly site in the yeast Saccharomyces cerevisiae. We have determined the three-dimensional reconstruction of the full S. cerevisiae Atg17-Atg31-Atg29 complex by single-particle electron microscopy. Our structure shows that Atg17-Atg31-Atg29 is dimeric and adopts a relatively rigid and extended "S-shape" architecture with an end-to-end distance of approximately 345 Å. Subunit mapping analysis indicated that Atg17 mediates dimerization and generates a central rod-like scaffold, while Atg31 and Atg29 form two globular domains that are tethered to the concave sides of the scaffold at the terminal regions. Finally, our observation that Atg17 adopts multiple conformations in the absence of Atg31 and Atg29 suggests that the two smaller components play key roles in defining and maintaining the distinct curvature of the ternary complex.
Keywords: Atg17; Atg29; Atg31; autophagy; single-particle electron microscopy.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures

References
-
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67. http://dx.doi.org/10.1038/nrm2708. - DOI - PubMed
-
- Kraft C, Martens S. Mechanisms and regulation of autophagosome formation. Curr Opin Cell Biol. 2012;24:496–501. http://dx.doi.org/10.1016/j.ceb.2012.05.001. - DOI - PubMed
-
- Noda NN, Ohsumi Y, Inagaki F. ATG systems from the protein structural point of view. Chem Rev. 2009;109:1587–98. http://dx.doi.org/10.1021/cr800459r. - DOI - PubMed
-
- Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–6. http://dx.doi.org/10.1016/j.febslet.2010.02.001. - DOI - PubMed
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. http://dx.doi.org/10.1146/annurev-cellbio-092910-154005. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
