MiR-26b is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion
- PMID: 23939832
- PMCID: PMC4030585
- DOI: 10.1002/path.4248
MiR-26b is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion
Abstract
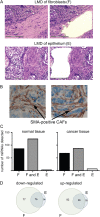
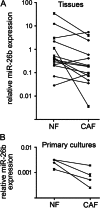
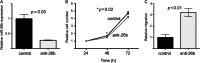
Carcinoma-associated fibroblasts (CAFs) influence the behaviour of cancer cells but the roles of microRNAs in this interaction are unknown. We report microRNAs that are differentially expressed between breast normal fibroblasts and CAFs of oestrogen receptor-positive cancers, and explore the influences of one of these, miR-26b, on breast cancer biology. We identified differentially expressed microRNAs by expression profiling of clinical samples and a tissue culture model: miR-26b was the most highly deregulated microRNA. Using qPCR, miR-26b was confirmed as down-regulated in fibroblasts from 15 of 18 further breast cancers. Next, we examined whether manipulation of miR-26b expression changed breast fibroblast behaviour. Reduced miR-26b expression caused fibroblast migration and invasion to increase by up to three-fold in scratch-closure and trans-well assays. Furthermore, in co-culture with MCF7 breast cancer epithelial cells, fibroblasts with reduced miR-26b expression enhanced both MCF7 migration in trans-well assays and MCF7 invasion from three-dimensional spheroids by up to five-fold. Mass spectrometry was used to identify expression changes associated with the reduction of miR-26b expression in fibroblasts. Pathway analyses of differentially expressed proteins revealed that glycolysis/TCA cycle and cytoskeletal regulation by Rho GTPases are downstream of miR-26b. In addition, three novel miR-26b targets were identified (TNKS1BP1, CPSF7, COL12A1) and the expression of each in cancer stroma was shown to be significantly associated with breast cancer recurrence. MiR-26b in breast CAFs is a potent regulator of cancer behaviour in oestrogen receptor-positive cancers, and we have identified key genes and molecular pathways that act downstream of miR-26b in CAFs.
Keywords: fibroblast; microRNA; microRNA-26b; stroma; tumour microenvironment.
© 2013 The Authors. Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Figures




References
-
- Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev. 1996;76:69–125. - PubMed
-
- Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nature Med. 2008;14:518–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

