Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin
- PMID: 23939888
- PMCID: PMC3811244
- DOI: 10.1128/AAC.01069-13
Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin
Abstract
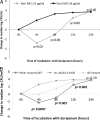
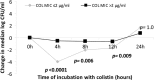
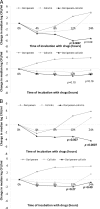
Doripenem-colistin exerts synergy against some, but not all, Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae strains in vitro. We determined if doripenem MICs and/or ompK36 porin gene mutations impacted the responses of 23 sequence type 258 (ST258), KPC-2-producing strains to the combination of doripenem (8 μg/ml) and colistin (2 μg/ml) during time-kill assays. The median doripenem and colistin MICs were 32 and 4 μg/ml. Doripenem MICs did not correlate with KPC-2 expression levels. Five and 18 strains had wild-type and mutant ompK36, respectively. The most common mutations were IS5 promoter insertions (n = 7) and insertions encoding glycine and aspartic acid at amino acid (aa) positions 134 and 135 (ins aa134-135 GD; n = 8), which were associated with higher doripenem MICs than other mutations or wild-type ompK36 (all P values ≤ 0.04). Bactericidal activity (24 h) was achieved by doripenem-colistin against 12%, 43%, and 75% of ins aa134-135 GD, IS5, and wild-type/other mutants, respectively (P = 0.04). Doripenem-colistin was more active in time-kill studies than colistin at 12 and 24 h if the doripenem MIC was ≤8 μg/ml (P = 0.0007 and 0.09, respectively), but not if the MIC was >8 μg/ml (P = 0.10 and 0.16). Likewise, doripenem-colistin was more active at 12 and 24 h against the wild type/other mutants than ins aa134-135 GD or IS5 mutants (P = 0.007 and 0.0007). By multivariate analysis, the absence of ins aa134-135 GD or IS5 mutations was the only independent predictor of doripenem-colistin responses at 24 h (P = 0.002). In conclusion, ompK36 genotypes identified ST258 KPC-K. pneumoniae strains that were most likely to respond to doripenem-colistin.
Figures

References
-
- Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430–1435 - PubMed
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 - PubMed
-
- Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 - PubMed
-
- Maltezou HC, Giakkoupi P, Maragos A, Bolikas M, Raftopoulos V, Paphatzaki H, Vrouhos G, Liakou V, Vatopouos AC. 2009. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece). J. Infect. 58:213–219 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

